Research Keywords
Antimicrobial/antifungal resistance, biophysics, machine learning, magnetobiology, mathematical/quantitative/computational/systems/synthetic biology, microbial evolution, nongenetic variability, pathogenic yeasts, synthetic gene circuits.
Antimicrobial Resistance
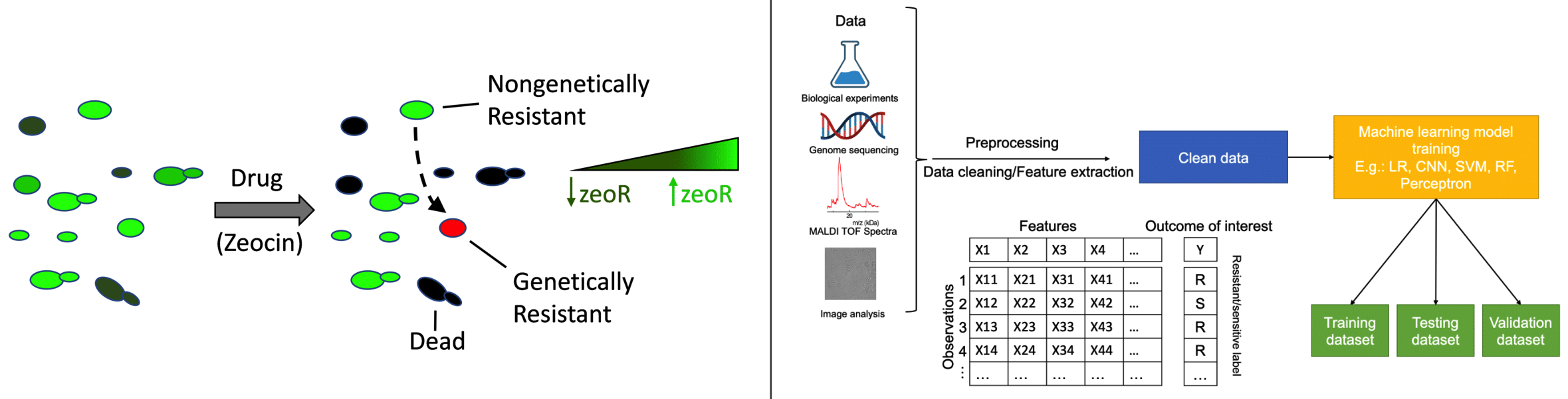
Antimicrobial resistance threatens the advances of modern medicine and has staggering global socio-economic costs. Pathogenic fungi are especially concerning due limited classes of antifungal drugs, unique challenges of antifungal drug development, and thermotolerance and species range expansion associated with climate change. We study antifungal resistance using physics-based models and by performing experiments on budding yeast (Saccharomyces cerevisiae) habouring synthetic gene networks and pathogenic fungi (Candida species). We also train machine learning models to quickly and accurately identify drug-resistant pathogens. This research uncovers novel forms of antimicrobial resistance and discovers new treatments against drug-resistant infections.
Selected Publications:
Peering inside the black box: Explainable AI to interpret advanced computer vision fungal pathogen prediction.
Guthrie, Shankarnarayan, and Charlebois. bioRxiv, doi: 10.1101/2025.06.27.662051 (2025).
Machine learning to identify clinically relevant Candida yeast species.
Shankarnarayan and Charlebois. Med. Mycol., 62: myad134 (2024).
Identification and elimination of antifungal tolerance in Candida auris.
Rasouli Koohi, Shankarnarayan, Galon, and Charlebois. Biomedicines, 11: 898 (2023).
Non-genetic resistance facilitates survival while hindering the evolution of drug resistance due to intraspecific competition.
Guthrie and Charlebois. Phys. Biol., 19: 066002 (2022).
Does transcriptional heterogeneity facilitate the development of genetic drug resistance?
Farquhar, Rasouli Koohi, and Charlebois. Bioessays, 43: 2100043 (2021).
Magnetobiology
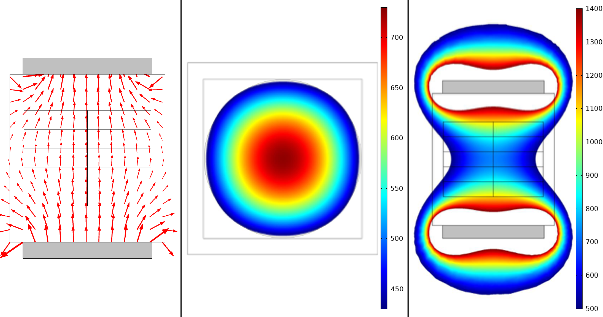
Magnetic fields have been shown to affect sensing, migration, and navigation in living organisms. However, the effects of magnetic fields on microorganisms largely remain to be elucidated. We develop spatiotemporal algorithms, build magnetic field devices, and perform experiments on yeast to study the effects of magnetic fields on cellular growth, gene expression, and fungal mat formation. This research generates open-source devices for magnetobiological research, advances our fundamental understanding of magnetic phenomena in microbes, and opens new possibilities for mitigating microbial biofilms.
Publications:
Magnetic field platform for experiments on well-mixed and spatially structured microbial populations.
Bandara, Li, and Charlebois. Biophys. Rep., 4: 100165 (2024).
Lattice-based Monte Carlo simulation of the effects of nutrient concentration and magnetic field exposure on yeast colony growth and morphology.
Hall and Charlebois. In Silico Biol., 14: 53-69 (2021).