J Pharm Pharmaceut Sci
(www.cspscanada.org) 9(1):101-112, 2006
Contribution of Polymorphisms in UDP-Glucuronosyltransferase and CYP2D6
to the Individual Variation in Disposition of Carvedilol
Yoh Takekuma1, Toru Takenaka2,
Masami Kiyokawa2, Koujiro Yamazaki2, Hiroshi Okamoto3,
Akira Kitabatake3, Hiroyuki Tsutsui3, Mitsuru Sugawara2
1Department of Clinical Pharmaceutics and Therapeutics,
Graduate School
of Pharmaceutical Science, Hokkaido University, Sapporo,
Japan
2Department of Pharmacy, Hokkaido
University Hospital,
Sapporo, Japan
3Department of Cardiovascular Medicine, Hokkaido University
Hospital, Sapporo, Japan
Received 17 December 2005, Revised 22 February 2006,
Accepted 23 February 2006, Published 1 March 2006.
Corresponding Author: Yoh Takekuma,
Department of Clinical Pharmaceutics and
Therapeutics, Graduate School of Pharmaceutical Science, Hokkaido University,
Kita-12-jo, Nishi-6-chome, Kita-ku, Sapporo 060-0812,
Japan.
y-kuma@pharm.hokudai.ac.jp
PDF
Version
ABSTRACT PURPOSE.
It has been reported that carvedilol, which has beta-adrenergic blocking and
vasodilating activities, is mainly metabolized by UDP-glucuronosyltransferase
(UGT) 1A1, UGT2B4, UGT2B7 and CYP2D6.
The aim of this study was to determine whether the activity of glucuronidation
has an influence on the area under the curve (AUC) of carvedilol and whether polymorphisms
in UGTs and CYP2D6 contribute to individual variation in disposition of
carvedilol in Japanese. METHODS.
Plasma concentrations of carvedilol and its glucuronide were determined by
reversed-phase high-performance liquid chromatography (HPLC). Genotyping of UGT1A1, UGT2B4 and UGT2B7 genes was carried out by the direct sequence method. CYP2D6 genotyping was carried out using
an amplification refractory mutation system (ARMS) assay and PCR-restriction
fragment length polymorphism (RFLP). RESULTS. The level of carvedilol glucuronidation ability in
the high-level AUC group was significantly lower than that in the low-level
group. The frequencies of UGT1A1*6, UGT2B7*3 and CYP2D6*10 in the low level ability of glucuronidation group were
significantly higher than those in the high level group, and the same tendency
was found in the frequency of CYP2D6*5,
though there was no significant difference. CONCLUSION. Polymorphisms of UGT1A1, UGT2B7 and CYP2D6 strongly affect the pharmacokinetics and disposition of
carvedilol in Japanese.
INTRODUCTION
Generally, orally
administered drugs are absorbed by the small intestine and then metabolized in
the liver. Metabolism includes phase I (oxidation, reduction, hydrolysis, etc.)
and phase II (conjugation) reactions. The phase I reaction introduces a
functional group such as a hydroxyl group onto the molecule or exposes a pre-existing
functional group, and the phase II reaction connects the functional group to an
endogenous species such as a glucuronic acid. Modified drug molecules are
hydrophilic and are excreted into bile and urine. However, some drugs do not
undergo the phase I reaction and are conjugated directly. It is possible that
individual variations in enzyme activity for conjugation affect the pharmacokinetics
of these drugs.
Carvedilol ((±)-1-carbazol-4-yloxy)-3-[[2-(omethoxyphenoxy)ethyl]-amino]-2-propanol) has badrenergic blocking and vasodilating activities [1, 2].
This drug is used to treat angina pectoris and hypertension and has recently
been used to treat chronic heart failure (CHF). However, for treatment of CHF,
it is recommended that the dose of carvedilol should be gradually and carefully
increased because of its negative inotropic activity [2-5].
It has been reported that carvedilol is metabolized by both
oxidation and conjugation pathways in the liver into various metabolites and
that the main pathway is direct glucuronidation of carvedilol because the main
metabolite in plasma and urine was found to be the glucuronide of unchanged
carvedilol (22% and 32%, respectively) [6, 7]. Three UDP-glucuronosyltransferase
(UGT) isoforms have been reported to be capable of conjugating carvedilol into
two forms of its glucuronides (G1 and G2) [8]. UGT2B4 formed both glucuronides,
whereas UGT1A1 (G2) and UGT2B7 (G1) formed either one. On the other hand,
oxidation pathways are mainly catalyzed by CYP2D6 [9]. CYP2D6 is responsible
for the formation of 4-hydroxy carvedilol and 5-hydroxy carvedilol, and both
metabolites are excreted into urine (6.4%) [7]. Therefore, we should not
disregard the influence of CYP2D6 in discussing the disposition of carvedilol, although
glucuronidation is the major metabolic pathway of carvedilol in humans.
The aim of this study was to clarify whether polymorphisms in UDP-Glucuronosyltransferase and CYP2D6 contribute to individual
variation in disposition of carvedilol.
MATERIALS AND METHODS
The study protocol was
approved by the Ethics Committee of the Graduate School of Medicine, Hokkaido
University. Written informed consent for participation in the study was obtained
from all subjects.
Chemicals
and regents
(±)
-Carvedilol was kindly supplied by Daiichi Pharmaceutical Co. (Tokyo, Japan).
Dihydroergotamine was obtained from Sigma-Aldrich (St. Louis, MO).
b-glucuronidase was purchased from Wako Pure Chemicals (Osaka, Japan).
All other reagents were of the highest grade available.
Patients and blood sampling
Forty-six patients (8
females and 38 males; median age, 65.5 (26-83) years; median body weight, 61.4
(32.0-98.9) kg) with CHF or angina pectoris who were being treated with
carvedilol were enrolled in this study. The patients with CHF were classified
into New York Heart Association (NYHA) class II-III. The daily doses of
carvedilol ranged from 1.25 to 20 mg, and the drug was taken in one or two
doses daily. The median creatinine clearance (Ccr) in the patients was 63.0 (15.9-156.6) mL/min. No patients had clinically overt
hepatic failure. There were no concomitantly used drugs that have been reported
to strongly influence plasma concentration of carvedilol. After a fixed dose of
carvedilol had been administered for 6 to 10 days, venous blood samples were
collected just before drug administration and at 1, 2, 4, 6, and 10 h after
administration.
Determination of carvedilol and its glucuronide
Plasma concentrations were
determined by reversed-phase high-performance liquid chromatography (HPLC) with
a fluorometric detector. The separation was performed on a GL-Pak Nucleosil
100-5C8 (4.6 mm I.D. x 250 mm) column (GL Science Inc., Tokyo, Japan).
The mobile phase was a mixture of acetonitrile and 50 mM potassium
dihydrogenphosphate (28:72) containing a final concentration of 5 mM
tetra-n-butylammonium chloride. The flow rate was 1.0 mL/min and column temperature
was 40șC. Excitation and emission wavelengths of 240 nm and 340 nm,
respectively, were used for fluorometric detection. For the determination of
unchanged carvedilol in plasma, 200 ”L of a sample was mixed with 2 mL of 1 M sodium hydroxide
and 200 mL of dihydroergotamine solution (20 mg/mL
in methanol) as an internal standard and then vortexed for 20 sec. Five mL of diethyl
ether was added to the mixture, and the mixture was shaken for 20 min. After centrifugation
at 1,800 x g for 5 min, the upper organic layer (4 mL) was evaporated to
dryness. The residue was reconstituted with 200 mL of
the mobile phase and 40 mL of them were injected into
the HPLC system. The lower limit of quantitation for carvedilol was 0.5 ng/mL. Coefficients of variation were 2.62% and 9.38% at
50 ng/mL and 0.5 ng/mL, respectively (n = 5). Determination of carvedilol
glucuronide was done after converting the glucuronide to parent carvedilol by b-glucuronidase;
that is, 200 mL of plasma was mixed with
20 mL of b-glucuronidase solution
(98,000 units/mL) and then incubated at 46șC for 1.5 h. After the reaction, total
carvedilol was determined as described above. The concentration of the
glucuronide was calculated by subtracting the concentration of the unchanged
form from the total concentration. The area under the curve (AUC0-10)
was calculated from the plasma concentration of carvedilol or its glucuronide
using the linear trapezoidal rule.
Genotyping
We obtained written informed
consent from 40 of 66 patients and genotyped their UGTs and CYP. Genomic DNA
was prepared using standard methods. The exons of UGT1A1, UGT2B4 and UGT2B7
genes (containing the promoter region of UGT1A1)
in 40 patients from whom written informed consents were obtained for genotyping
were sequenced.
Each exon was amplified from genomic DNA (20-60 ng) using 0.5
units of Ex-Taq (Takara Bio Inc., Shiga,
Japan) with 1 mM
of the primers shown in
Table 1.
The conditions of polymerase
chain reaction (PCR) were as follows: denaturation at 94șC for 5 min, followed
by 30 cycles comprising denaturation at 94șC for 30 sec, annealing at 54-64șC
for 10 to 30 sec, and extension at 72șC for 30 to 80 sec, and then a final
extension at 72șC for 7 min. The PCR products were treated by ExoSap-IT (Takara
Bio Inc.) at 37șC for 15 min and at 80șC for 15 min to degrade the excess
primers and dNTP. The products were directly sequenced with the primers listed
in Table 1 using a Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems,
Foster City, CA) according to the manufacturers
recommended protocol.
For CYP2D6 variants,
the same 40 patients were genotyped. Mutant alleles that have been reported to
have high frequencies in the Japanese population were selected in this study
[10]. Genotyping of CYP2D6*5 (deletion
of the CYP2D6 allele) was carried out
using an amplification refractory mutation system (ARMS) assay as described by
Johansson et al. [11] and Steen et al. [12] with minor modification. Genotyping
of 100C>T (common SNP to CYP2D6*4,
*10, *14, *36, *37, *47 and *49)
was carried out using the ARMS assay as described by Johansson et al. [13].
Then samples that had the 100T allele were genotyped on CYP2D6*4 and CYP2D6*14. Genotyping
of CYP2D6*4 was carried out using the
ARMS assay as described by Heim et al. [14]. Genotyping of CYP2D6*14 was carried out using the PCR-restriction fragment length
polymorphism (RFLP) assay as described by Wang et al. [15]. In this study,
subjects with 100C>T mutation were classified into CYP2D6*10 except for CYP2D6*4
and *14 because frequencies of CYP2D6*18, *21, *36, *37, *47 and *49 are rare in the Japanese population
[10].
Statistical analysis
Data are expressed as mean ± SD.
Differences in the ability of glucuronidation between low and high level AUC
groups were measured using students t-test. Differences between allele frequencies
in the two groups were measured using Fishers exact test. Correlation between
the numbers of mutant alleles and the ability of glucuronidation was analyzed
using Spearmans rank correlation test. A p
value below 0.05 was considered statistically significant.
RESULTS
Pharmacokinetics property of carvedilol
Plasma concentrations of
carvedilol and its glucuronide in the 46 patients were determined, and 66
profiles of AUC0-10 were obtained. Pharmacokinetic parameters are
listed in Table 2.
Table
2.
Pharmacokinetic
parameters of carvedilol in tested patients.
|
Groups of usage
|
Normalized AUC
(hr/kL)
|
Cmax/dose
(kL-1)
|
tmax
(h)
|
t1/2
(h)
|
|
Once a day
(n = 17)
|
17.8±22.6
|
3.35±3.79
|
3.23±1.02
|
2.99±1.30
|
|
Twice a day
(n = 49)
|
28.7±23.8
|
4.95±4.20
|
3.49±1.95
|
6.32±5.75
|
Normalized
AUC: ratio of AUC0-10 to dose of carvedilol.
Data are given as
means±SD.
No difference was found
between the tested subjects and Japanese healthy subjects in tmax
and t1/2 [16]. However, ratios of AUC0-10 to dose of
carvedilol (normalized AUC) and Cmax/dose in the subjects were
higher than those in Japanese healthy subjects [16].
Normalized AUCs of each subject are shown in
Figure 1.
The range of normalized AUCs was very wide in both the once a
day and twice a day group (2.69-85.1 and 2.83-108.9, respectively) and it was
independent of dose. It was ascertained whether individual variation of ability
to glucuronidate was responsible for this individual variation of normalized
AUCs. In general, the ratio of metabolite to parent drug AUC is used as
indicator of metabolization ability. However, in this study, we defined the
metabolic index (MI) as follows because clearance of carvedilol glucuronide
depends on renal function:
MI = (AUC glucuronized x Ccr) / AUC unchanged, (1)
where AUC glucuronized and AUC
unchanged are AUC of glucuronized carvedilol and AUC of unchanged carvedilol, respectively
and Ccr is creatinine clearance.
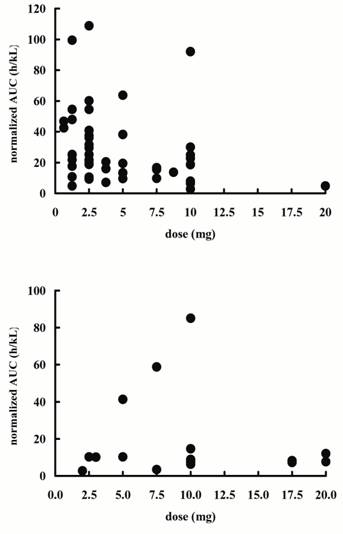
Figure 1: Distribution of ratios of AUC0-10
to dose of carvedilol (normalized AUC) at each dose in the patients taking
carvedilol. The top graph is the group of
twice a day (n=49) and bottom graph is the group of once a day (n=17).
The subjects were classified
into low and high level normalized AUC groups, and the metabolic indexes in
these two groups were compared (Figure 2).
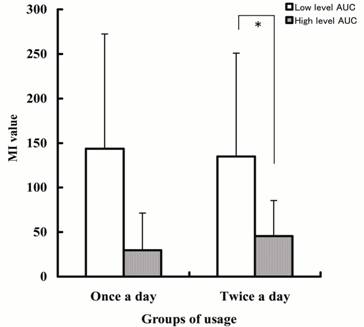
Figure 2: Comparison of the ability of
carvedilol glucoronidation in the low and high level AUC groups. MI value: ratio of carvedilol glucuronide to
unchanged carvedilol AUCs which was multiplied by creatinine clearance. Each
column represents the mean with SD. *; p<0.001.
In the twice a day group, MI
value was significantly lower in the high level AUC group than in the low level
AUC group (p<0.01). In the once a day group, the same
tendency was found, but though there was no significant difference.
Genotyping
Genotyping of UGT1A1, UGT2B4, and UGT2B7 in the 40 patients that gave written informed consent for genotyping
was carried out. For reference sequences, AF297093 was used for UGT1A1, and NT_077444.2 (GenBank
accession numbers) was used for UGT2B4
and UGT2B7.
Table 3
shows the results of genotyping. The subjects were
classified into low and high level MI groups. DNA sequence analysis confirmed
the presence of variants of UGT1A1*6
(211G>T, G71R), UGT1A1*28 (A (TA)
6TAA to A (TA) 7TAA), UGT2B7*2
(802C>T, H268Y) and UGT2B7*3
(211G>T, A71S) in UGT2B7. In UGT2B4, no SNP was found except for a silent
mutation (1212A>T).
Frequencies of UGT1A1
and UGT2B7 alleles in the 40 subjects
are shown in
Table 4. No
significant differences were found between low and high level MI groups with UGT2B7*2. However, the frequencies of UGT1A1*6, UGT1A1*28 and UGT2B7*3
were significantly different between the two groups. The frequencies of UGT1A1*6 and UGT2B7*3 in the low level MI group were higher than those in the
high level MI group, but the frequency of UGT1A1*28
in the low level MI group was lower than that in the high level MI group.
As shown in Tables 3 and 4, genotyping for four CYP2D6 alleles was carried out in the
same 40 patients genotyped for UGT.
Variants of CYP2D6*4, *5 and *10 were detected. Only one subject had the CYP2D6*4 allele. The frequency of CYP2D6*10 in the low level MI group was two-times higher than that
in the high level group, and the
same tendency was found in the frequency of CYP2D6*5,
though there was no significant difference.
Figure 3 shows the relation between numbers of mutant alleles
except for UGT2B7*2 and MI values.
One CYP2D6*5 or *4 allele is counted as two alleles because both alleles lack
catalytic activity and it is thought that influence of those alleles on
metabolic activity is greater than that of the other alleles. MI values showed
a tendency to decrease with increase in the number of mutant alleles. (p<0.001).
DISCUSSION
This is the first study to
demonstrate the relationship between polymorphisms of UGTs and CYP2D6 and
disposition of carvedilol at the same time. Our results indicated large
variations in the normalized AUC of carvedilol. These variations were found in
all groups of patients receiving carvedilol of doses of 1.25 mg to 10 mg
(Figure 1).
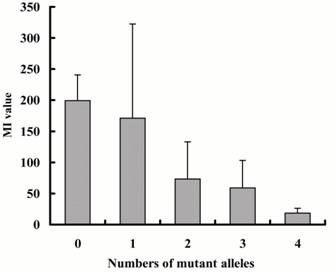
Figure 3: Relationship between numbers of
mutant alleles except for UGT2B7*2
and ability of carvedilol glucoronidation. One
CYP2D6*5 or *4 allele is counted as two alleles because both alleles lack
catalytic activity. M1 values: ratio of carvedilol glucoronide to unchanged
carvedilol AUCs which was multiplied by creatinine clearance. Each column
represents the mean with SD. Significant correlation by Spearmans rank
correlation (p<0.001).
None of the patients
receiving doses of more than 15 mg had particularly high AUC. Since the dose of
carvedilol in patients who showed reductions in blood pressure and heart rate
was not increased, patients administered such a high dose of carvedilol may not
a show high plasma concentration. Normalized AUC in the present study
(2.69-85.1 hr/kL in the once a day group) was much higher than that in healthy
adults (2.56-15.0 hr/kL) reported by Neugebauer et al. [6]. It has been
reported that plasma concentrations of carvedilol were increased in patients
with CHF compared with concentrations in healthy volunteers (50-100% higher
values in patients with NYHA class Ⅳ CHF) [17]. The increase in plasma concentration is thought
to be caused by a reduction in uptake of carvedilol to the liver accompanied by
a decrease in the bloodstream.
However, this cannot account for our results because some of
the AUC values in the present study were six-times higher than those in healthy
adults, whereas patients with CHF show values only 3-4 times higher than those
in healthy adults.
Figure 2 shows that MI, an indicator of the ability of
glucuronidation of carvedilol, was associated with normalized AUC. In twice a
day group, the MI value was significantly lower in the high level AUC group
than in the low level AUC group (p<0.01). In once a day group, the same
tendency was found, though there was no significant difference because the
number of subjects was too small. These results suggest that the ability of
glucuronidation affects the AUC of carvedilol, in accordance with results of
previous studies showing that the main metabolic pathway is glucuronidation of
carvedilol [6, 7].
Ohno et al. [8] showed by using a recombinant UGT assay that UGT1A1,
UGT2B4 and UGT2B7 are responsible for glucuronidation of carvedilol. Therefore,
these three UGT isoforms were sequenced in the 40 patients to determine whether
polymorphisms of these genes are responsible for the variation in the ability
of glucuronidation of carvedilol. No missense mutation in UGT2B4 was found in the 40 patients (Table 3).
Only a few mutations in UGT2B4,
namely, UGT2B4*2 (1374T>A, D458E),
*3 (325T>T and 1186T>C, F109L
and F369L), *4 (1364A>G, K455R)
and *5 (1531T>C, C511R), have been
reported [18-20]. The frequencies of the UGT2B4*2
allele in Caucasian and African populations have been reported to be 20.0% and
15.0%, respectively, by Lampe et al. [21] and Riedy et al. [22]. However, this
mutation in Japanese is rare [20]. Our results are in agreement with those
reports. In UGT1A1, many variant
alleles have been reported (http://som.flinders.edu.au/FUSA/ClinPharm/UGT/).
In this study, UGT1A1*6 (211G>A,
G71R) and *28 (A (TA) 6TAA to A (TA)
7TAA) were detected. Of the 40 patients in this study, the genotypes of UGT1A1*6 were homozygous in 1 patient and
heterozygous in 13 patients. All of them except for one patient were in the low
level MI group (Table 3).
The frequency of this mutation in Japanese has been reported to
be 13-16% [23, 24]. Yamamoto et al. [25] reported that the catalytic activity
level of the UGT1A1 enzyme was
reduced to 30% in subjects with a homozygote for the UGT1A1*6 allele. Therefore, UGT1A1*6
was thought to reduce the activity of carvedilol glucuronidation. The allele
frequency of UGT1A1*28 in the 40
patients was 13.7%. This finding is in agreement with results of previous
studies [24, 26, 27]. However, the allele frequency of
UGT1A1*28 in the high level MI group
was higher than that in the low level MI group (Table 4).
UGT1A1*28 has been
reported to be associated with a 20-80% reduction in gene expression [28, 29].
Our results do not reflect these reports. On the other hand, it has reported
that a homozygote for UGT1A1*28
reduced the activity of estradiol glucuronidation in microsomes from the human
liver to 23%, whereas a heterozygote reduced the ability to 82% [30]. All of
the subjects with UGT1A1*28 in this
study were heterozygous. Therefore, it is thought that a heterozygote for UGT1A1*28 had little effect of
carvedilol glucuronidation. As for UGT2B7,
UGT2B7*2 (802C>T, H268Y) and *3
(211G>T, A71S) were found in this study (Tables 3 and 4). UGT2B7*2 was the most frequently found
variant allele in Japanese in previous studies [20, 31]. Although no remarkable
functional difference between UGT2B7*1 and
UGT2B7*2 alleles was found in several
studies [32-35], one study has shown that subjects who had a UGT2B7*2 allele showed a significantly
higher morphine-6-O-glucuronide / morphine ratio than did subjects with UGT2B7*1 [36]. Our results showed that
there was no significant difference between the low and high level MI groups in
the allele frequency of UGT2B7*2
(Table 4). On the other hand, the frequency of UGT2B7*3 in the low level MI group was significantly higher than
that in the high level group. UGT2B7*3
has been reported by Hirota et al. [31] and Saeki et al. [20]. The effect of
this allele on catalytic activity is still unknown. Our results suggest that UGT2B7*3 allele reduces the activity of carvedilol
glucuronidation. Therefore, it is possible that UGT1A1*6 and UGT2B7*3 are
responsible for the low level of glucuronidation activity of carvedilol.
With regard to CYP2D6 as an oxidative enzyme, the frequencies
of CYP2D6*4, *5 and *10 in the 40 patients were 1.25%, 5.0%
and 36.3%, respectively. These results are in agreement with results of
previous study [10]. The frequency of CYP2D6*10 in the low level MI group was two-times higher than that
in the high level group, and the
same tendency was found in the frequency of CYP2D6*5,
though there was no significant difference.
The AUC of R (+)-carvedilol in patients who were poor metabolizers
of debrisoquin (an indicator of low level of CYP2D6 activity) was 2.56-times
higher than that of R (+)-carvedilol in patients who were extensive
metabolizers of debrisoquin. In contrast, the AUC of S (-)-carvedilol in poor
metabolizers of debrisoquin and that in extensive metabolizers of debrisoquin
were similar [37]. Honda et al. reported of effect of CYP2D6*10 on the pharmacokinetics of R- and S-carvedilol in healthy
Japanese [38]. Accordingly, it is necessary to take polymorphisms of CYP2D6 into consideration when investigating
the pharmacokinetics and disposition of carvedilol.
Although MI is an indicator of carvedilol glucuronidation
activity, the frequencies of variant alleles that affect the oxidative
catalytic activity of CYP2D6 were different in the low and high level MI
groups. One possible reason for this is that reduction of catalytic activity of
CYP2D6 leads to an increase in the unchanged carvedilol plasma concentration
and AUC as the denominator of MI. On the other hand, the absolute quantity of
carvedilol glucuronides does not greatly change. As a result, the MI value as an
indicator of glucuronidation ability of carvedilol decreases. It is possible
that MI is a good indicator of total metabolic activity of carvedilol including
UGT and CYP2D6.
Figure 3 shows the relation between numbers of mutant alleles
except for UGT2B7*2 and MI values.
One CYP2D6*5 allele is counted as two
alleles because the CYP2D6*5 allele
is a whole deletion of the CYP2D6*5
gene and it is thought that the influence of this allele on metabolic activity
is greater than that of the other alleles. MI values showed a tendency to
decrease with increase in the number of mutant alleles (p<0.001), indicating
that polymorphisms of UGT1A1, UGT2B7
and CYP2D6 affect carvedilol disposition
in cooperation.
Giessmann reported that CYP2D6
genotype and intestinal expression of P-glycoprotein (P-gp) and
multidrug-resistant protein 2 (MRP2) are the major variables in carvedilol
disposition [39]. Our result is agreement with effect of CYP2D6 genotype. However, polymorphism for UGT should be taken into
consideration because carvedilol glucuronides are major metabolites. In one
mutant allele group, the range of MI values was very wide compared with that in
the other groups. The contribution of other metabolic pathways and intestinal
expression of P-gp or MRP2 in carvedilol absorption are possible to be the
reason for this. Our results demonstrated that individual variations in the
disposition of carvedilol, which is metabolized by multiple pathways, are
caused not only by the polymorphism for the main enzyme, UGT, but also by another enzymes such as CYP2D6.
Conclusions
Forty patients who were
being treated with carvedilol were phenotyped and genotyped for UGT1A1, UGT2B4, UGT2B7 and CYP2D6.
The allele frequencies of UGT1A1*6,
UGT2B7*3 and CYP2D6*10 in the low
level MI group were significantly higher than those in the low level MI group.
MI values showed a tendency to decrease with increase in the number of mutant
alleles. These results suggest that polymorphisms of UGT1A1, UGT2B7 and CYP2D6
strongly affect the pharmacokinetics and disposition of carvedilol.
References
[1] von Mollendorff E, Abshagen U, Akpan W, Neugebauer G, Schroter
E. Clinical pharmacologic investigations with carvedilol, a new beta-blocker
with direct vasodilator activity. Clin Pharmacol Ther 1986, 39: 677-82
[2]
Frishman WH. Carvedilol. N Engl J Med 1998, 339: 1759-65
[3]
Packer M, Bristow MR, Cohn JN, Colucci WS, Fowler MB,
Gilbert EM, Shusterman NH. The effect of carvedilol on morbidity and mortality
in patients with chronic heart failure. U.S. Carvedilol Heart Failure Study
Group. N Engl J Med 1996, 334: 1349-55
[4]
Cice G, Ferrara L, D'Andrea A, D'Isa S, Di Benedetto A,
Cittadini A, Russo PE, Golino P,Calabro R. Carvedilol increases two-year
survival in dialysis patients with dilated cardiomyopathy: a prospective,
placebo-controlled trial. J Am Coll Cardiol 2003, 41: 1438-44
[5]
Keating GM, Jarvis B. Carvedilol: a review of its use in
chronic heart failure. Drugs 2003, 63: 1697-741
[6]
Neugebauer G, Akpan W, von Mollendorff E, Neubert P, Reiff
K. Pharmacokinetics and disposition of carvedilol in humans. J Cardiovasc
Pharmacol 1987, 10 Suppl 11: S85-8
[7]
Neugebauer G, Neubert P. Metabolism of carvedilol in man.
Eur J Drug Metab Pharmacokinet 1991, 16: 257-60
[8]
Ohno A, Saito Y, Hanioka N, Jinno H, Saeki M, Ando M, Ozawa
S, Sawada J. Involvement of human hepatic UGT1A1, UGT2B4, and UGT2B7 in the
glucuronidation of carvedilol. Drug Metab Dispos 2004, 32: 235-9
[9]
Oldham HG, Clarke
SE. In vitro identification of the human cytochrome P450 enzymes involved in
the metabolism of R(+)- and S(-)-carvedilol. Drug
Metab Dispos 1997, 25: 970-7
[10]
Soyama A, Kubo T, Miyajima A, Saito Y, Shiseki K, Komamura
K, Ueno K, Kamakura S, Kitakaze M, Tomoike H, Ozawa S, Sawada J. Novel nonsynonymous
single nucleotide polymorphisms in the CYP2D6 gene. Drug Metab Pharmacokinet
2004, 19: 313-9
[11]
Johansson I, Oscarson M, Yue QY, Bertilsson L, Sjoqvist F, Ingelman-Sundberg
M. Genetic analysis of the Chinese cytochrome P4502D locus: characterization of
variant CYP2D6 genes present in subjects with diminished capacity for
debrisoquine hydroxylation. Mol Pharmacol 1994, 46: 452-9
[12]
Steen VM, Andreassen OA, Daly
AK, Tefre T, Borresen AL, Idle JR,Gulbrandsen AK. Detection of the poor
metabolizer-associated CYP2D6(D) gene deletion
allele by long-PCR technology. Pharmacogenetics 1995, 5: 215-23
[13]
Johansson I, Lundqvist E, Dahl ML, Ingelman-Sundberg M.
PCR-based genotyping for duplicated and deleted CYP2D6 genes. Pharmacogenetics
1996, 6: 351-5
[14]
Heim M,Meyer UA. Genotyping of
poor metabolisers of debrisoquine by allele-specific PCR amplification. Lancet
1990, 336: 529-32
[15]
Wang SL, Lai MD, Huang JD. G169R mutation diminishes the
metabolic activity of CYP2D6 in Chinese. Drug Metab Dispos 1999, 27: 385-8
[16]
Fujimaki M, Hakusui H, Hasegawa Y, Ajima H, Ota H, Igafashi
S,Yamamura H. Pharmacokinetics of Carvedilol (DQ-2466) in Healthy Subjects. Jpn
J Clin Pharmacol Ther 1990, 21: 415-424
[17]
Carlson W, Oberg K. Clinical Pharmacology of Carvedilol. J
Cardiovasc Pharmacol Ther 1999, 4: 205-218
[18]
Jin C, Miners JO, Lillywhite KJ, Mackenzie PI.
Complementary deoxyribonucleic acid cloning and expression of a human liver
uridine diphosphate-glucuronosyltransferase glucuronidating carboxylic
acid-containing drugs. J Pharmacol Exp Ther 1993, 264: 475-9
[19]
Levesque E, Beaulieu M, Hum DW, Belanger A.
Characterization and substrate specificity of UGT2B4 (E458): a
UDP-glucuronosyltransferase encoded by a polymorphic gene. Pharmacogenetics
1999, 9: 207-16
[20]
Saeki M, Saito Y, Jinno H, Tanaka-Kagawa T, Ohno A, Ozawa
S, Ueno K, Kamakura S, Kamatani N, Komamura K, Kitakaze M, Sawada J. Single
nucleotide polymorphisms and haplotype frequencies of UGT2B4 and UGT2B7 in a
Japanese population. Drug Metab Dispos 2004, 32: 1048-54
[21]
Lampe JW, Bigler J, Bush AC, Potter JD. Prevalence of
polymorphisms in the human UDP-glucuronosyltransferase 2B family: UGT2B4(D458E), UGT2B7(H268Y), and UGT2B15(D85Y). Cancer
Epidemiol Biomarkers Prev 2000, 9: 329-33
[22]
Riedy M, Wang JY, Miller AP, Buckler A,
Hall J, Guida M. Genomic organization of the UGT2b gene cluster on human
chromosome 4q13. Pharmacogenetics 2000, 10: 251-60
[23]
Akaba K, Kimura T, Sasaki A, Tanabe S, Ikegami T, Hashimoto
M, Umeda H, Yoshida H, Umetsu K, Chiba H, Yuasa I, Hayasaka K. Neonatal
hyperbilirubinemia and mutation of the bilirubin uridine
diphosphate-glucuronosyltransferase gene: a common missense mutation among
Japanese, Koreans and Chinese. Biochem Mol Biol Int 1998, 46: 21-6
[24]
Maruo Y, Nishizawa K, Sato H, Doida Y, Shimada M.
Association of neonatal hyperbilirubinemia with bilirubin
UDP-glucuronosyltransferase polymorphism. Pediatrics 1999, 103: 1224-7
[25]
Yamamoto K, Sato H, Fujiyama Y, Doida Y, Bamba T.
Contribution of two missense mutations (G71R and Y486D) of the bilirubin UDP
glycosyltransferase (UGT1A1) gene to phenotypes of Gilbert's syndrome and
Crigler-Najjar syndrome type II. Biochim Biophys Acta 1998, 1406: 267-73
[26]
Ando Y, Chida M, Nakayama K, Saka H, Kamataki T. The
UGT1A1*28 allele is relatively rare in a Japanese population. Pharmacogenetics
1998, 8: 357-60
[27]
Hall D, Ybazeta G, Destro-Bisol G, Petzl-Erler ML, Di
Rienzo A. Variability at the uridine diphosphate glucuronosyltransferase 1A1
promoter in human populations and primates. Pharmacogenetics 1999, 9: 591-9
[28]
Beutler E, Gelbart T, Demina A. Racial variability in the
UDP-glucuronosyltransferase 1 (UGT1A1) promoter: a balanced polymorphism for
regulation of bilirubin metabolism? Proc Natl Acad Sci U S A 1998, 95: 8170-4
[29]
Bosma PJ, Chowdhury JR, Bakker C, Gantla S, de Boer A,
Oostra BA, Lindhout D, Tytgat GN, Jansen PL, Oude Elferink RP, et al. The
genetic basis of the reduced expression of bilirubin
UDP-glucuronosyltransferase 1 in Gilbert's syndrome. N Engl J Med 1995, 333:
1171-5
[30]
Fisher MB, Vandenbranden M, Findlay K, Burchell B, Thummel
KE, Hall SD, Wrighton SA. Tissue distribution and interindividual variation in
human UDP-glucuronosyltransferase activity: relationship between UGT1A1
promoter genotype and variability in a liver bank. Pharmacogenetics 2000, 10:
727-39
[31]
Hirota T, Ieiri I, Takane H, Sano H, Kawamoto K, Aono H,
Yamasaki A, Takeuchi H, Masada M, Shimizu E, Higuchi S, Otsubo K. Sequence
variability and candidate gene analysis in two cancer patients with complex
clinical outcomes during morphine therapy. Drug Metab Dispos 2003, 31: 677-80
[32]
Bhasker CR, McKinnon W, Stone A, Lo AC, Kubota T, Ishizaki
T, Miners JO. Genetic polymorphism of UDP-glucuronosyltransferase 2B7 (UGT2B7)
at amino acid 268: ethnic diversity of alleles and potential clinical
significance. Pharmacogenetics 2000, 10: 679-85
[33]
Holthe M, Klepstad P, Zahlsen K, Borchgrevink PC, Hagen L,
Dale O, Kaasa S, Krokan HE, Skorpen F. Morphine glucuronide-to-morphine plasma
ratios are unaffected by the UGT2B7 H268Y and UGT1A1*28 polymorphisms in cancer
patients on chronic morphine therapy. Eur J Clin Pharmacol 2002, 58: 353-6
[34]
Holthe M, Rakvag TN, Klepstad P, Idle JR, Kaasa S, Krokan
HE, Skorpen F. Sequence variations in the UDP-glucuronosyltransferase 2B7
(UGT2B7) gene: identification of 10 novel single nucleotide polymorphisms
(SNPs) and analysis of their relevance to morphine glucuronidation in cancer
patients. Pharmacogenomics J 2003, 3: 17-26
[35]
Court MH, Krishnaswamy S, Hao Q, Duan SX, Patten CJ, Von
Moltke LL, Greenblatt DJ. Evaluation of 3'-azido-3'-deoxythymidine, morphine,
and codeine as probe substrates for UDP-glucuronosyltransferase 2B7 (UGT2B7) in
human liver microsomes: specificity and influence of the UGT2B7*2 polymorphism.
Drug Metab Dispos 2003, 31: 1125-33
[36]
Sawyer MB, Innocenti F, Das S, Cheng C, Ramirez J,
Pantle-Fisher FH, Wright C, Badner J, Pei D, Boyett JM, Cook E, Jr., Ratain MJ.
A pharmacogenetic study of uridine diphosphate-glucuronosyltransferase 2B7 in
patients receiving morphine. Clin Pharmacol Ther 2003, 73: 566-74
[37]
Zhou HH,Wood AJ. Stereoselective
disposition of carvedilol is determined by CYP2D6. Clin Pharmacol Ther 1995,
57: 518-24
[38]
Honda M, Nozawa T, Igarashi N, Inoue H, Arakawa R, Ogura Y,
Okabe H, Taguchi M, Hashimoto Y. Effect of CYP2D6*10 on the pharmacokinetics of
R- and S-carvedilol in healthy Japanese volunteers. Biol Pharm Bull 2005, 28:
1476-9
[39]
Giessmann T, Modess C, Hecker U, Zschiesche M, Dazert P,
Kunert-Keil C, Warzok R, Engel G, Weitschies W, Cascorbi I, Kroemer HK, Siegmund
W. CYP2D6 genotype and induction of intestinal drug transporters by rifampin
predict presystemic clearance of carvedilol in healthy subjects. Clin Pharmacol Ther 2004, 75: 213-222
JPPS Contents
Published by the Canadian Society for
Pharmaceutical Sciences.
Copyright © 1998 by the Canadian
Society for Pharmaceutical Sciences.
http://www.cspscanada.org/
CSPS Home |
JPPS Home |
Search |
Subscribe to JPPS


