J Pharm Pharmaceut Sci
(www.cspscanada.org) 8(3):586-592, 2005
Quantitative Structure Antitumoral-Activity
Relationships of Thiadiazinthione Derivatives Using the Novel Hybrid Molecular
Index pMRc
Ramón
Carrasco,a Juan A. Padrón,b
Rolando Pérez,c Hortensia Rodríguez,c Margarita Suárez,c
Carmen Ochoad
aDept. Química,
Centro de Química
Farmacéutica, La Habana, Cuba
bLaboratorio de Química Computacional y
Teórica, Facultad de Química, Universidad de La Habana
cLaboratorio de Síntesis Orgánica, Facultad
de Química, Universidad de La Habana
dInstituto de Química Médica (CSIC)
Madrid, España
Received October 26, 2005; Revised November 18,
2005, Accepted November 25, 2003, Published December
1, 2005
Abstract
Purpose.
The recently defined
molar-refractivity-partition index was
applied to a family of 1,3,5-
thiadiazin-2-thione derivatives in
order to establish quantitative
structure-antitumoral models. The goal
of this effort is to establish the
relationships between the structure
and biological response of these
compounds.
Method. After the splitting of
the sample in two sets, their indices
were correlated against the measured
biological activity. The combined use
of our index with others had been able
to describe not only the topologic but
also the
London
dispersive forces of any fragment in
relation to the biological response of
the sets.
Results. The obtained
models showed correlation coefficients
of 0.87 and 0.81 respectively linking
structural and biological features of
the molecules. The mean relative error
values were less than 7%. According to
the models, the activity of the first
sample is related mostly to molecular
topology and dispersive forces. Sample
two activity was associated to the
size and branching of the
substituents, and also to the
London
forces.
Conclusion. The index was able
to discriminate between pure
topological features and those related
to dispersive forces.
Introduction
3,5-disubstituted-tetrahydro-2H-1,3,5-thiadiazin-2-thione derivatives
(I) have a wide spectrum of
antimicrobial activity.[1] Several studies related to the antifungic, antiviral,
antihelmintic, and tuberculostatic activity of these compounds have been
extensively reported.[1-3] Previously, two series of 3,5-disubstituted
thiadiazinthiones have been synthesized and assayed in vitro and in vivo in
order to obtain new antiparasitic chemical substances for assessment as potential
antiprotozoal agents.[2] In a recent work the antitumoral activity of these
compounds have been reported against several tumoral cell lines and also some
features of the degradation pathways of this heterocycle[3] were outlined. Up to now, experimental evidence strongly suggests that the
thiadiazinthione ring acts as a prodrug. [1] In a protic medium, the
heterocycle undergoes a ring opening to form different active metabolites,
which are indeed responsible of the biological activity [1]. The use of
topologic and topographical indexes is widely accepted for molecular modeling.
Recently, we reported the hybrid molar-refractivity-partition index (pMRc)[4,5],
based on Randic algorithm [6] and Ghose and Crippen partitioned molecular
refractivity [7], that have the capability to portray London dispersive forces
from a different perspective than the molecular refractivity. Then, the aim of
this work is to test the index capability to relate structural features of this
compound family with their antitumoral activity.
Theoretical Details
Molar
Refractivity
The molar refractivity is a
constitutive-additive property calculated by the Lorenz-Lorentz formula:
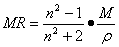 (1)
(1)
where M is the molecular weight, n
is the refraction index and r the density, and its value depends only of the wavelength of the
light used to measure the refraction index. For a radiation of infinite
wavelength, the molar refractivity represents the real volume of the molecules
and its polarizability. Then, the molar refractivity is
related, not only to the volume of the molecules but also to the London dispersive forces
that act in the drug-receptor interaction.
The atomic contribution to molecular refractivity
calculated by Ghose and Crippen method
Ghose and Crippen defined 110
atom types, [7] representing most commonly occurring atomic states of carbon,
hydrogen, oxygen, nitrogen, halogens, and sulphur in organic molecules to split
the molar refractivity. They stated that this classification partially
differentiates the polarizing effects of heteroatoms and the effect of
overlapping with non-hydrogen atoms, although they accepted that this
classification might be weak in differentiating the conjugation effects. The
authors stated that the classification may not completely cover all organic molecules,
and that addition of atom types is always feasible. Further on, the 110 atom
types were reduced to 22 (table 2). They assumed that the sum of the atomic
values (ai) is the
molecular value of the molar refractivity (eq. 2):
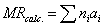 (2)
(2)
Graph theory. The Randic-type graph theoretical invariant
Graph theory is a branch of
mathematics related to topology and combinatorial problems.[18]
Different authors have reported a large number of topological and topographical
indices as well as its broad and successful applicability in QSAR and QSPR
studies. However, the principal problems of the use of this approach are related to the physical meaning and the information
redundancy among indices of similar definition, commonly expressed by high
correlation values. [19] In this sense, Randic postulated [20] that any novel
molecular descriptor 1) need to be simple, 2) add more insights to the problem,
3) or to solve a unexplained problem by alternative schemes, among others
features. Randic molecular connectivity index for path of order p ( pc) is
defined as [6]:
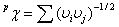 (3)
(3)
It was modified by Kier and Hall[21,22] by
defining subgraphs Gj tree type in the G graph containing edges. To
each vertex i of graph G is
associated a term
di (for example di = vi). After this, to each subgraph Gi
with vertices j1,..., jh+1 is calculated the F
magnitude (Eq. 4)
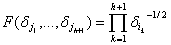 (4)
(4)
The numbers F(di1, dj(h+1)) are then added in all the subgraph
Gi.. To include the multiple bonds and heteroatoms, Kier and Hall suggested the
employ of di as,
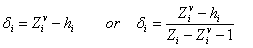 (5)
(5)
where Zi is the
total number of electrons, Zvi
is the number of valence electrons and hi
is the hydrogen atoms number bonded to atom i. The connectivity indices
obtained by this formula are known as valence indices [22]
and represented by pcv. A more detailed explanation of the
graph theory and calculation procedures can be found elsewhere.[23]
Molar refractivity partition index
pMRc
Molar refractivity partition index[6,7] (pMRc) use the Randic-type graph-theoretical invariant, and is defined as
follow (Eq. 6),
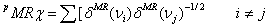 (6)
(6)
where the sum is all over adjacent vertices in the graph. dMR(vi) is the atomic refractivity value of the
vi atom. The atomic refractivity values of
bonded hydrogen to heavy atoms are also added to this
term, to take into account their contribution in the graph.
Experimental part
Table 1
shows the compound
sets. Reported antitumoral activity [3] is shown in
tables 3 and 5. Biological activity is given as log IC50 against HeLa cells (logA). All structures were optimized to
0.01 convergence by the semiempirical method PM3 [9,10]
implemented in MOPAC 6.0 program. [11-13]
In all the indexes employed
the superscript p and c indicates path and cluster order, respectively.
Randić (pc) and valence (pccV), based on the vertices connectivity matrix and pe Estrada indices, in the edge connectivity
matrix [14] were used as topological descriptors. As topographic
descriptors, the pW, pWq, pWqC and per Estrada indices [14] were employed. The W indices are based in the matrix of the
vertices, weighted by the bond orders, the charge density and the charge
density with spatial correction respectively. In the case of per, the connectivity matrix of the edges is weighted
with the bond orders. The c, cV, W, Wq, WqC,
e and er
were calculated with the MODEST program
[15]. Both the
bond orders and the charge density were taken from the
output of the semiempirical calculations.
pMRc index was included to portray the importance of the London dispersive forces.
Paths of order 1 to 6, clusters of order 3 and 4, and combinations of cluster 3
with path order 1 to 3 were calculated, using a program developed in the
authors laboratory [16] The log P and
MR values were calculated by the Ghose and Crippen methodology.
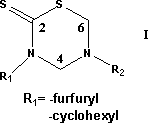
The 27 compounds in the sample were divided in two, according the nature of R1
substituent. Forward stepwise multiple regression analysis was
employed to establish the quantitative regression models, using the
STATISTICA package. [17] A statistical outlier was defined
as any compound that failed in three of the several criteria included in the
program. In such case it was excluded from the sample
and the analysis was restarted. Both models were cross-validated,
reporting the Q2 value.
Results and Discussion
The mechanism of the
antitumoral action of these compounds is not well established. It has been previously described that, in biological
environment, the thiadiazinthione ring breaks to generate active metabolites.
Since only two different substituents in position 1 are
represented in the series, it is possible to use an indicator (dummy) variable
(taking values 1 or 0 for the presence or not of a given substituent) to
described the whole set. However, we had preferred to treat the sample
separately. Therefore the sample was divided in two:
Sample #1 with R1=furfuryl and Sample #2 with R1=cyclohexyl.
Analysis of the sample set
#1
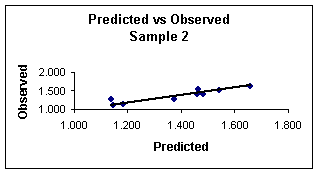
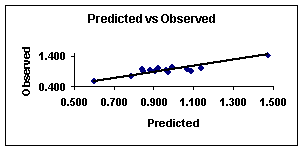
Figure 3.
Predicted
vs. observed values calculated with equation 8 for sample 2.
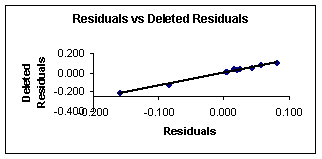
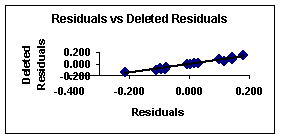
Figure 4. Residuals vs. deleted
residuals obtained from equation 8.
Figures
3
and
4 show the plots of the predicted vs. observed values with equation 8, and
the graph of residuals vs. deleted residuals.
This last graph (fig. 4) suggests that this regression model is
more stable than that given with equation 7 for the
first compounds set. This can be explained from the
definition of deleted residuals. They are the residuals that one would obtain
if the respective case would be excluded from the estimation of the multiple regression (i.e., the computation of the regression
coefficients).
Thus, if there are large discrepancies between the deleted
residuals and the regular standardized residuals, then it can be concluded that
the regression coefficients are not very stable, that is, they are greatly
affected by the exclusion of single cases. If these results are
analyzed in the same manner as for sample 1, it may be concluded that,
qualitatively speaking, 90% of the sample was correctly predicted.
Conclusion
As a general remark, when the
substituent in R1 is furfuryl, an increased activity is obtained except when R
is CH[(CH2)2COOH]-COOH. Equations
7 and 8 suggest that the presence of path-cluster fragments type (2MR3c and 3MR3c respectively) in both series also increases the activity. These two
features suggest that this pattern of substitution affords leads compounds in
both series. On the other hand, it was determined that the pMRc index is
useful to describe the antitumoral activity of the thiadiazinthiones set
studied in this work. The index was able to discriminate between pure
topological features and those related to dispersive forces.
Acknowledgements
This work has
been partially supported by Ministry of Public Health of Cuba grants.
The authors wish to thanks the referees and the editorial board for their
useful comments and kindly patient.
References
[1] Ertan, M.; Bilgil, A.A.; Palaska, E. and
Yulug, N.; Synthesis and Antifungal Activities of some 3-(2-Phenylethyl)-5-substituted-tetrahydro-2-1,3,5-thiadiazine-2-thione,
Arzneimitttel Forschung-Drug Research,
42:160-171, 1992.
[2] a) Ochoa, C.; Pérez, E.; Pérez, R.;
Suárez, M.; Ochoa, E.; Rodríguez, H.; Barrio, A.G.; Muelas, S.; Nogal, J.J. and
Martínez, R.A.; Synthesis and Antiprotozoan Properties of New 3,5-Disubstituted-Tetrahydro-2H-1,3,5-Thiadiazin-2-Thione
Derivatives. Arzneimitttel Forschung-Drug
Research, 49:(II), 464-469, 1999. b) Muelas, S.;
Suárez, M.; Pérez, R.; Rodríguez, H.; Ochoa, C.; Escario, J.A. and Gómez-Barrio, A.; In Vitro and In Vivo Assays of 3,5-Disubstituted Tetrahydro-2H-1,3,5-thiadiazin-2-thione Derivatives
Against Trypanosoma cruzi. Mem. Inst.
Oswaldo Cruz, Río de Janeiro, 97:(2), 269-272, 2002.
[3] Pérez, R.; Rodríguez,
H.; Pérez, E.; Suárez, M.; Reyes, O.; González, L.J.; López de Cerain, A.; Ezpeleta, O.; Pérez, C. and Ochoa. C.; Study on the Decomposition Products of
Thiadiazinthione and their Anti-Cancer Properties. Arzneim. Forsch. -Drug Res.; 50(II): 854-857,
2000.
[4] Padrón, J.A.; Carrasco, R. and Pellón,
R.; Definition of New Descriptor Based on Molar Refractivity Partition Using
Randic Type Graph-Theoretical Invariant.; Revista Cubana de Farmacia, 34 Special Supp. Especial June
2000 pp. 328-2.
[5] Padrón, J. A.; Carrasco, R. and Pellón,
R.F.; Molecular Descriptor Based on a Molar Refractivity Partition using
Randic-type Graph-theoretical Invariant,
J.Pharm.Pharmaceut. Sci.(www.ualberta.ca/~csps)
5(3):234-231, 2002.
[6] Randic, M.; On Characterization of
Molecular Branching, J. Am. Chem. Soc.,
97:6609-6615, 1975.
[7] a) Ghose, A.K.
and Crippen, G.M.; Atomic Physicochemical Parameters for Three-dimensional
Structure-Directed Quantitative Structure-Activity Relationships. I. Partition
Coefficients as a Measure of Hydrophobicity. J.Comput.
Chem., 7:(4), 565-577, 1986; b) Ghose, A.K. and
Crippen, G.M.; Atomic Physicochemical Parameters for Three-Dimensional
Structure-Directed Quantitative Structure-Activity Relationships. 2. Modeling Dispersive and Hydrophobic Interactions; J. Comput. Chem.; 27(1):21-35, 1987.
[8] HyperChem,
(1993) Hypercube, Inc. and Autodesk, Inc. Release 3 for Windows.
[9] Stewart, J.J.P.; Optimization of
Parameters for Semi-empirical Methods I-Method; J.Comp.Chem., 10:209-220, 1989.
[10] Stewart, J.J.P.; Optimization of Parameters
for Semi-empirical Methods II-Applications;
J.Am.Chem.Soc. 10:221-264, 1989.
[11] Stewart, J. J. P.; MOPAC 6.0. Indiana University, Bloomington, IN,
Manual QCPE # 581.
[12] Stewart, J. J. P.; Mopac: A
Semiempirical Molecular Orbital Program, J. Comp. Aid.
Mol. Design, 4(1): 1-103, 1990.
[13] MOPAC
v.6 for 3/486/Pentium PC’s. Release 1.02; March 1997; Windows 95 and NT
environments. Universidad de La Habana, Cuba, 1994-97.
[14] a) Estrada, E. and Montero, L.A.;
Bond Order Weighted Graphs in Molecules as Structure-property Indices. Mol. Eng., 2:363-373, 1993. b) Estrada, E.
and Ramírez, A.; Edge Adjacency Relationships and Molecular Topographic
Descriptors. Definition and QSAR Applications. J.
Chem. Inf. Comput. Sci., 37:837-843, 1997;
c) Estrada,
E. Three-dimensional Molecular Descriptors Based on Electron Charge Density
Weighted Graphs. J. Chem. Inf. Comput. Sci. 35:708-713, 1995; d) Estrada, E.; Edge Adjacency Relationships and a Novel Topological
Index Related to Molecular Volume. J. Chem. Inf.
Comput. Sci., 35:31-33, 1995; e) Estrada, E. and Molina,
E.; 3D Connectivity Indices in QSPR/QSAR Studies. J. Chem. Inf. Comput. Sci. 41:791-797, 2001.
[15] Rodríguez, L.; Estrada,
E.; Muñoz, I. and Gutiérrez Y.; MODEST (Molecular DESign Tools) 2.0.
Universidad Central de Las Villas: Santa Clara, 1994.
[16] Padrón, J.A. and Carrasco, R.; La Habana,
Program for the calculation of pMRc Index. Pharmaceutical Chemistry Center, 2002.
[17] STATISTICA
for WINDOWS, version 4.0, Statsoft, Inc. 1993.
[18] Harary,
F.; Graph Theory, Addison-Wesley, Reading
MA, 1969.
[19] Basak,
S.C.; Magnuson, V.R.; Niemi,
G.J.; Regal, R.R. and Veith, G.D.; Topological Indices: Their Nature, Mutual
Relatedness, and Applications, Math.
Model, 8:300-305, 1987.
[20] Kubinyi, H., QSAR: Hansch Analysis and
Related Approaches, pp. 59-60. In: Methods and Principles in Medicinal
Chemistry, R. Mannhold, P. Krogsgaard-Larsen y H. Timmerman, Eds. 1993,
[19] Golbraikh, A. and Tropsha, A. J. Mol. Graph. Model., 20, 269-276(2002).
[20] Randic, M.; Hansen, P.J. and Jurs, P.C.;
Search for Useful Graph Theoretical Invariants of Molecular Structure, J. Chem. Inf. Comput.Sci., 28:60-68,
1988.
[21] Kier, L.B. and Hall,
L.H.; Molecular Connectivity in Chemistry
and Drugs Research, Academic Press, New
York, 1976.
[22] Kier, L.B. and Hall,
L.H.; Molecular Connectivity in
Structure-Activity Analysis; Research Studies Press Ltd., Hertfordshire,
England and John Wiley and
Sons, New York
1986.
[23] Molconn-Z 3.50 Manual;
http://www.eslc.vabiotech.com/molconn/manuals/350/
JPPS Contents
Published by the Canadian Society for
Pharmaceutical Sciences.
Copyright © 1998 by the Canadian
Society for Pharmaceutical Sciences.
http://www.cspscanada.org/
CSPS Home |
JPPS Home |
Search |
Subscribe to JPPS
![]() (1)
(1)
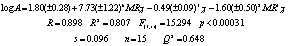 (7)
(7)
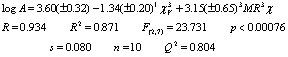 (8)
(8)
