J Pharm Pharmaceut Sci (www.cspscanada.org) 8(2):348-360, 2005
Structural
toxicity relationship of 4-alkoxyphenols’ cytotoxicity towards murine B16-F0
melanoma cell line
Majid Y.
Moridani1, Mike Moore2, Richard A. Bartsch3,
Yanfei Yang3, Souzan Heibati-Sadati3
1Department
of Pharmaceutical Sciences, School of Pharmacy; Department of Pediatrics, School
of Medicine, Texas Tech University
HSC, Amarillo, Texas, USA
2School of
Pharmacy, Texas Tech University HSC, Amarillo, Texas,
USA
3Department
of Chemistry and Biochemistry, Texas Tech University,
Lubbock,
Texas, USA
Received May 18 2005, Revised June 23 2005,
Accepted June 26 2005, Published August 18 2005
PDF
Version
ABSTRACT. PURPOSE. The aim of this study was to identify phenolic
agents that could form quinone reactive intermediate metabolites in melanocytes
in order to be effective as anti-melanoma agents; but were not metabolized by
liver P450 metabolizing enzymes in order to have minimal toxicity towards the
liver. METHODS. Tyrosinase, an
enzyme present abundantly in melanocytes was selected as a molecular target for
the treatment of malignant melanoma. Ten alkoxyphenols were investigated for
their metabolism by tyrosinase/O2, rat liver P450 microsomal/NADPH/O2
metabolizing systems and for their toxicity towards B16-F0 melanoma cells. RESULTS. All the alkoxyphenols showed a
dose- and time-dependent toxicity towards B16-F0 cells except
2-iso-propoxyphenol. 4-n-Hexyloxyphenol demonstrated the greatest toxicity
towards B16-F0 cells while minimally depleting glutathione in microsomal
preparations at its calculated LC10 and LC50 lethal concentrations
for B16-F0. At 100 mM concentrations,
4-t-butoxyphenol showed the lowest amount of glutathione depletion by
microsomal P450 system. Alkoxyphenols with at least two alkyl groups derivatized
at alpha carbon of alkoxy group showed minimal rates of metabolism by tyrosinase/O2
metabolizing system. A quantitative structural toxicity relationship equation
was also derived, LogLC50(mM)=
–0.265(±0.064)LogP + 2.482(±0.179). CONCLUSIONS.
4-n-hexyloxy-phenol was identified as a potential lead anti-melanoma agent against
B16-F0 melanoma cells with minimal metabolism by rat liver P450 microsomal
preparation.
INTRODUCTION
Malignant melanoma is one of the deadliest cancers
known to man. It is estimated that 55,100 new invasive melanoma cases are diagnosed
in the USA
every year of which 7,910 will be expected to die from the disease (1). The
estimated lifetime risk for melanoma among Americans is 1 in 74. Currently, the
therapy for melanoma includes surgical intervention, which has a high rate of
treatment failure in highly metastatic and advanced cases that are usually
fatal. Systemic chemotherapy is
often the only resource, but the results to date have been very disappointing
and the lack of selective cytotoxicity often leads to intolerable side effects (2).
With increasing occurrence of this disease, there is a clear and urgent need
for an improved treatment regimen with enhanced specificity.
Tyrosinase, an enzyme found abundantly only in
melanocytes, was selected as a molecular target for 4-hydroxyanisole (4-HA) in
the past. 4-HA is a simple phenolic agent which was first shown by Riley (3) to
be a melanocytotoxic agent. Tyrosinase was shown to catalyze the oxidation of
4-HA to 4-methoxycatechol and its o-quinone,
which reacted readily with nucleophiles (4–6). In addition, melanoma toxicity
may result from the covalent binding of the o-quinone to protein thiols and/or glutathione (GSH) depletion (7)
and inhibition of mitochondrial electron transport (8). This ultimately leads
to desirable melanoma cell death. 4-HA was the only compound from this class
that was tested in clinical trials as an anti-melanoma agent (3, 9). Depigmentation
and tumor shrinkage resulted from both the topical application of 4-HA (3) and
intra-arterial infusions of 4-HA into patients’ legs (9). Unfortunately, 4-HA clinical
trails were terminated because serious liver damage occurred (10) but there
were no insights into the mechanisms resulting in induced liver toxicity. It
was recently shown that 4-HA was also metabolized by liver P450s via arene
epoxidation route to p-quinone (Figure 1), a reactive metabolite, which was highly toxic to isolated rat hepatocytes (6).

Figure 1: Metabolism pathway
for 4-HA in melanocyte and hepatocyte. 4-HA was metabolized by liver P450s via arene
epoxidation route to p-quinone, a reactive metabolite, which can deplete GSH by
conjugate formation. P-Quinone was shown to be highly toxic to isolated rat hepatocytes (6). 4-HA was also metabolized
by melanocyte tyrosinase to form 4-methoxycatechol and then an o-quinone which
can react with intracellular GSH and is toxic towards melanoma cells.
In the current work, we sought to
identify a phenolic compound with minimum toxicity towards the liver but yet efficacious
against melanoma. We thus investigated ten alkoxyphenol compounds with various
linear aliphatic side chains and their branched analogues (Figure 2) for their
metabolism by tyrosinase/O2, rat liver P450 microsomal preparation/NADPH/O2
metabolizing systems, and for their toxicity towards the B16-F0 mouse melanoma
cell line. Our data indicates that all alkoxyphenols tested in this work demonstrated
toxicity towards murine B16-F0 melanoma cell line. It was postulated that only
4-nHP (4-n-hexyloxyphenol) demonstrated a significant advantage over other
alkoxyphenols with respect to GSH depletion by rat liver microsomal P450s and therefore
its toxicity towards the liver.
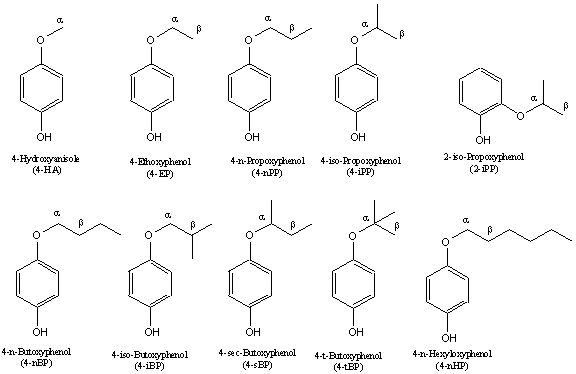
Figure 2: Chemical structure
of alkoxyphenols. The
positions of hydrogen and the side chains are marked as a and b on the aliphatic side chain of alkoxyphenols.
EXPERIMENTAL SECTION
Materials
All materials, solvents and reagents were purchased from
either Sigma-Aldrich, USA,
or Fisher-Scientific, USA. DETAPAC (diethylenetriaminopentaacetic
acid) was purchased from Sigma-Aldrich,
USA. Mushroom
tyrosinase was used throughout this study. Phenolic agents that were investigated
in this study included: 4-HA (4-hydorxyanisole); 4-EP (4-ethoxyphenol); 4-nPP
(4-n-propoxyphenol); 4-iPP (4-iso-propoxyphenol); 2-iPP (2-iso-propoxyphenol);
4-nBP (4-n-butoxyphenol); 4-iBP (4-iso-butoxyphenol); 4-sBP
(4-sec-butoxyphenol); 4-tBP (4-t-butoxyphenol); and 4-nHP (4-n-hexyloxyphenol).
4-t-Butoxyphenol was purchased from Matrix
Scientific, USA.
4-iPP, 4-iBP and 4-sBP were synthesized (11). Dulbecco’s Modified Eagle Medium
(DMEM) (Cat. No. 11965-092), fetal bovine serum (Cat. No. 10082-139),
Penicillin-Streptomycin; (10,000 units/mL, Cat. No. 15140-122) and Versene
(1:5000 Cat. No. 15040-066) were purchased from Invitrogen, USA.
The mouse melanoma B16-F0 cell line was obtained from American Type Culture
Collection (ATCC), USA.
Methods
Syntheses of 4-iso-propoxyphenol,
4-sio-butoxyphenol and 4-sec-butoxyphenol
All alkoxyphenols were commercially available except
4-iso-butoxyphenol, 4-sec-butoxyphenol and 4-iso-propoxyphenol, which were
prepared by adaptation of a method published by Naish-Byfield et al (11). Briefly,
KOH (0.020 mol) dissolved in ethanol (80 mL) was added dropwise over a 1 h
period to a stirred solution of hydroquinone (0.10 mol) and the appropriate
alkyl bromide (0.020 mol) in ethanol (400 mL) at reflux under nitrogen. After
overnight refluxing and cooling to room temperature, excess base was
neutralized with acetic acid. The inorganic salts were filtered and the solvent
was evaporated in vacuo to give a
mixture of hydroquinone and the 4-alkoxyphenol. Dichloromethane was added and
the mixture was filtered to remove most of the excess hydroquinone. The
filtrate was dried over magnesium sulfate and evaporated in vacuo. The crude product was purified by recrystallization or
column chromatography. 4-iso-Butoxyphenol was obtained in 50% yield after
recrystallization from dichloromethane-hexane (mp 53-64oC). IR
(deposit on a NaCl plate from dichloromethane solution): 3359 (OH), 1235 (CO),
1039 (CO) cm-1. 1H NMR (300 MHz, CDCl3): d = 1.01 (d, J = 6.6 Hz, 6H, CH3), 1.99-2.07 (m, 1H, CH), 3.66 (d, J = 6.6 Hz, 2H, CH2), 4.37
(s, 1H,
OH), 6.76-6.77 (m, 4H, ArH).
4-sec-Butoxyphenol was isolated in 30%
yield after chromatography on silica gel. IR (film): 3359 (OH), 1230 (CO), 1100
(CO) cm-1. 1H NMR (300 MHz, CDCl3): d = 0.98 (t, J = 7.2 Hz, 3H, CH3CH2),
1.25 (d, J = 6.0 Hz, 3H, CH3 CH), 1.47-1.82 (m, 2H, CH2),
4.08-4.12 (m, 1H, CH), 4.59 (s, 1H, OH),
6.75-6.77 (m, 4H, ArH). Analysis, Calculated for C10H14O2:
C, 72.26; H, 8.44. Found: C, 71.86; H, 8.45.
4-iso-Propoxyphenol was isolated in 50%
yield after chromatography on silica gel. IR (film): 3359 (OH), 1226 (CO), 1120
(CO) cm-1. d = 1.29
(d, J = 6.0 Hz, 6H, CH3),
4.38-4.42 (m, 1H, CH), 4.69 (s, 1H, OH),
6.75-6.77 (m, 4H, ArH). Analysis, Calculated for C9H12O2:
C, 71.03; H, 7.95. Found: C, 71.29; H, 7.90.
UV-VIS spectroscopy of tyrosinase
mediated metabolism of alkoxyphenols
The spectra of a solution containing alkoxyphenol (100
mM) and tyrosinase (20
U/mL) were recorded in the absence and presence of GSH (200 mM) using a GBC UV-Visible spectral
spectrophotometer (GC Scientific,
Australia). The
spectra of the mixture were obtained when GSH was added to the solution either
before or after the addition of tyrosinase. The control spectrum was that of the
respective alkoxyphenol solution (100 mM) in phosphate buffer [0.1 M (pH 7.4) containing
DETAPAC (1 mM)].
Tyrosinase mediated GSH depletion
assay
Tyrosinase
(10 mL;
2500 U/mL) was added to a mixture of alkoxyphenol (100 mM) and GSH (200 mM) in a final volume of 1 mL phosphate
buffer (0.1 M, pH 7.4, DETAPAC 1mM). The mixture was pre-incubated for 30, 90,
and 180 min at 37°C. A 250 mL aliquot was added to trichloroacetic acid (25 mL; 30% w/v), vortexed and left at room
temperature for 5 min. A 100 mL aliquot of the supernatant was then added to a
mixture of Ellman’s reagent 5,5’-dithiobis-(2-nitrobenzoic acid) DTNB (25 mL; 2 mg/mL) and Tris/HCl buffer (875 mL; 0.1 M, pH 8.94), and then vortexed. The
absorbance of the solution was observed at 412 nm (12, 13). The standard curve
for GSH measurement gave a regression coefficient of greater than 0.99 over the
range of 5-200 mM GSH concentrations (data not shown).
Animal housing and protocol
Adult male Sprague-Dawley rats, 250–300g, were
obtained from Charles River Laboratories, USA, fed ad libitum, were allowed to
acclimatize for 1 week on clay chip bedding in a room with a 12 h light photocycle,
an environmental temperature of 21-23 °C and 50-60% relative humidity. The animal
protocols used in current investigation for rat liver microsomal preparation were
reviewed and approved by Institutional Animal Care and Use Committee at Texas
Tech University Health Sciences Center, Amarillo, TX.
Rat liver microsomal preparation
The rats were
anesthetized by sodium pentobarbital (60 mg/kg) before surgery in order to
prepare the animal before liver removal.
Hepatic microsomes were prepared by differential centrifugation as
described previously (14). Briefly, the liver was removed and weighed in a
beaker on ice. The liver was cut into pieces and washed by cold KCl (154 mM):
Tris/HCl (50 mM, pH 7.4) buffer solution then suspended into 4 volumes of
KCl:Tris/HCl buffer. The tissue was gently homogenized using an electrical
homogenizer and subsequently by a handheld glass tissue grinder before
centrifuging at 1935 g (Beckman Avanti J-25I, Beckman Rotor- JA-25.5) at 4 °C
for 15 min to remove tissue and cell debris. The supernatant was centrifuged at
12,100 g at 4 °C for 15 min to remove subcellular organelles followed by
centrifugation at 100,000 g (Beckman Optima LE-80K, Beckman Rotor- 45 Ti) at 4 °C
for 1 h. The supernatant was discarded and micorsomes were separated and
suspended in 5 mL Tris/HCl buffer (100 mM, pH 7.4) containing 1 mM DETAPAC. The
mixture was homogenized using a handheld glass tissue grinder after which an additional
15 mL Tris/HCl buffer was added and the solution was aliquoted in 750 mL and stored at -70°C for subsequent use. Microsomal
protein content was determined by a modified Lowry method (15).
CYP2E1 induced rat liver microsomal
preparation
CYP2E1 induced microsomes
were prepared from rats treated (i.p.) with inducing agent pyrazole (200
mg/kg/day) (16) for 2 consecutive days before sacrificing the rats on the 3rd
day.
Microsomal mediated GSH depletion
assay
The amount of
GSH conjugates formed was determined colorimetrically using Ellman’s reagent
5,5’-dithiobis-(2-nitrobenzoic acid) (DTNB) (13). Incubation mixtures contained
in a final volume of 1 mL phosphate buffer (0.1 M, pH 7.4, DETAPAC 1mM), 2 mg/mL
rat liver microsomes, 200 mM GSH, 1
mM NADPH, and 100 mM alkoxyphenols. The
mixtures were gently mixed at 37 °C from which 250 mL aliquots were taken at 30, 60, and 90 min time-interval
points into Eppendorf tubes containing 25 mL trichloroacetic acid (30% w/v). Following protein
precipitation and centrifugation for 5 min, the GSH levels of a 100 mL aliquot of the supernatant was determined
by the addition of 0.1 M Tris//HCl buffer, pH 8.94 (875 mL), and 2 mg/mL DTNB (25 mL). The reduced DTNB formed was determined
at 412 nm on a GBC spectral spectrophotometer.
Cell Culture
The mouse B16-F0
cell line was obtained from American Type Culture Collection, USA. A frozen B16-F0 cell vial was
washed twice with DMEM media before culturing in 8 mL DMEM supplemented with
fetal bovine serum (FBS) (10%) and streptomycin/penicillin (100 U/mL) in a T-25
flask (17). All the cell culture processes were carried out in a type II
vertical laminar air flow chamber. The B16-F0 cultures were kept at 37 °C under
a 5% CO2 atmosphere in a TS Autoflow CO2 Water-Jacketed
Incubator (NUAIRE, USA). The culture medium was
changed when acidification was indicated by the pH indicator and when needed.
To detach the cells from the flask, the media was first removed from the flask.
The remaining media was then washed out using sterile phosphate buffered saline
(PBS) (2-3 mL). Subsequently, Versene (2-5 mL) was added to the flask and the
sample was incubated for 2–3 min in the incubator to provide time for cells to
detach. The detached cells were rinsed with ~10 mL of pre-warmed sterile PBS to
dilute Versene. The mixture was transferred into a 50 mL tube. The flask was additionally
rinsed with sterile PBS and the content was added to the rest of the cells collected.
The cells were then spun down at 800 RPM (Beckman GPR Centrifuge, USA) for
3-5 min. The PBS-Versene mixture was aspirated off. Pelletted cells were then re-suspended
in DMEM media (supplemented by FBS 10% and antibiotics 100 U/mL) followed by
splitting the mixture into one T-75 flask containing 30 mL media (25% of the
media was supplemented from the previous culture step as conditioning media).
Cell viability
To determine cell viability, the cells obtained from each
flask were suspended in 4 mL of DMEM media supplemented by FBS 10% and
antibiotics 100 U/mL (contained 25% media from the previous culture step as
conditioning media). The cells were counted using trypan blue exclusion method (18)
for determining the viability.
MTT assay in 96 well plates
To evaluate
cytotoxicity, cells were obtained from exponentially growing 90-95% confluent
cultures and seeded at 12,500 cells/well in 96-well plates. The cells were kept
in 100 mL fresh DMEM media (supplemented by FCS 10%
and antibiotics 100 U/mL) for 24 h to allow cell adhesion and environmental
adaptation. Subsequently, the cells were treated with additional 150 mL DMEM (supplemented with FCS 10% and
antibiotics 100 U/mL) containing various concentrations of alkoxyphenols for
1-4 days. At 24 h interval, the medium was removed and the wells were washed
three times using DMEM media alone before adding 40 mL of 2 mg/mL yellow tetrazolium dye (3 - ( 4, 5- dimethylthiazolyl
- 2 ) - 2, 5 - diphenyltetrazolium bromide) (MTT) (17). The plates were
returned to the incubator for a period of 4 h. The residual MTT solutions were
removed from wells and then 200 mL of DMSO
was added to each well. The plates were stored at room temperature in a dark
place for an additional 2 h before reading them at 570 nm using XFluor Plate
Reader (Tecan US, Inc, USA). All experiments were performed in triplicate. An
analysis of variance (ANOVA) of repeated means was carried out to compare the
percentage of surviving cells in the cultures for different concentrations of
each compound. t-test was used to compare the results of toxicity of the alkoxyphenols
with 4-HA.
LC10 and LC50 calculation
Lethal
concentrations (LC) which can cause 10% and 50% of the cell death were
calculated from the linear regression equation derived from graphing the
viability of the cells at day 2 (on x axis) versus the concentration of the
drug (on y axis).
Partition coefficient
Partition
coefficient values were estimated using the LogP software available at
www.LogP.com.
Table 1: Metabolism mediated percentage GSH depletion
of alkoxyphenols by tyrosinase and rat liver microsomal preparations
|
|
B16-F0 toxicity (mM)
(day 2)
|
|
Metabolism mediated %GSH depletion
|
|
Alkoxyphenols
|
Eq. constructed from
data (day 2)a
|
Observed LCb
|
Estimatedc
LC50 (Eq.2)
|
LogPd
|
Tyrosinase/O2e
|
Normal liver
microsomes/
NADPH/O2f
|
2E1 induced liver
microsomes/
NADPH/O2f
|
|
|
|
LC10
|
LC50
|
Log
LC50
|
LC50
|
Log
LC50
|
|
30
min
|
90
min
|
180
min
|
90
min
|
30
min
|
60
min
|
90
min
|
|
4‑hydroxyanisole
|
LC= - 1.2 viability (%) + 118
|
10
|
58**
|
1.76
|
117
|
2.07
|
1.56
|
87±3†
|
100
|
100**
|
10±3
|
30±3
|
42±3
|
58±2**
|
|
4‑ethoxyphenol
|
LC= - 1.4 viability (%) + 145
|
18
|
74
|
1.87
|
90
|
1.95
|
2.00
|
7±1
|
43±2
|
87±3
|
11±3
|
27±3
|
43±2
|
52±5
|
|
4‑n‑propoxyphenol
|
LC= - 1.5 viability (%) + 132
|
<1
|
58
|
1.76
|
68
|
1.83
|
2.45
|
4±1
|
54±5
|
87±4
|
23±4
|
39±4
|
52±3
|
61±3
|
|
4‑iso‑propoxyphenol
|
LC= - 1.9 viability (%) + 181
|
14
|
88
|
1.94
|
72
|
1.86
|
2.35
|
2±1
|
9±2
|
13±2*
|
24±3
|
42±2
|
57±3
|
67±3
|
|
2-iso-propoxyphenol
|
LC= - 4.6 viability (%) + 484
|
70
|
254
|
2.40
|
75
|
1.88
|
2.29
|
1±1
|
9±2
|
17±2*
|
18±4
|
41±2
|
51±4
|
56±4
|
|
4-n-butoxyphenol
|
LC= - 1.0 viability (%) + 96
|
6
|
46
|
1.66
|
52
|
1.71
|
2.90
|
21±2
|
88±4
|
100
|
27±3
|
45±3
|
58±2
|
69±3
|
|
4-iso-butoxyphenol
|
LC= - 1.2 viability (%) + 134
|
24
|
74
|
1.87
|
55
|
1.74
|
2.81
|
49±3
|
87±4
|
100
|
32±4
|
47±3
|
73±3
|
75±3
|
|
4-sec-butoxyphenol
|
LC= - 1.1 viability (%) + 118
|
19
|
62
|
1.79
|
56
|
1.75
|
2.78
|
1±1
|
7±2
|
17±3*
|
25±3
|
42±4
|
60±4
|
66±3
|
|
4-t-butoxyphenol
|
LC= - 4.3 viability (%) + 414
|
27
|
200*
|
2.30
|
55
|
1.74
|
2.80
|
1±1
|
5±1
|
19±2*
|
33±5
|
8±3
|
19±1
|
25±3*
|
|
4-n-hexyloxyphenol
|
LC= - 1.8 viability (%) + 116
|
<1
|
26*
|
1.41
|
29
|
1.46
|
3.87
|
94±2†
|
100
|
100
|
38±3
|
42±4
|
60±4
|
66±3
|
a LC (lethal concentration)
equation was derived from regression analysis of viability data on the day 2
versus concentration.
b LC10 and LC50
were calculated from equations presented in the first column (see above).
c The values were calculated
from LogLC50 (mM)=
– 0.265LogP + 2.482 (QSTR Eq.2).
d LogP was calculated from LogP
software available from www.logP.com.
e The mixture contains
tyrosinase (25 units/mL), alkoxyphenol (100 mM) and GSH (200 mM)
in a final volume of 1 mL phosphate buffer (0.1 M, pH 7.4, DETAPAC 1mM).
f The reaction mixture contains
microsomes (2 mg/mL), alkoxyphenol (100 mM) and GSH (200 mM)
in a final volume of 1 mL phosphate buffer (0.1 M, pH 7.4, DETAPAC 1mM).
* Significantly different (t-test,
P<0.05) from 4-HA (marked as **) for selected data.
† As effective as 4-HA for being metabolized
by mushroom tyrosinase/O2 metabolizing system.
RESULTS
UV-VIS spectroscopy of tyrosinase
mediated metabolism of alkoxyphenols
The progression
of alkoxyphenol’s oxidation was monitored by tyrosinase/O2 oxidizing
system using a UV-VIS spectroscopy method, which showed a distinct peak at
420-470 nm with a characteristic indicative of o-quinone formation. Addition of
glutathione at the beginning of the metabolism reaction resulted in the
significant loss in the absorbance of the 420-470 nm peaks. The UV-VIS spectra
of this peak for all the alkoxyphenols were developed over 1 min except for
2-iPP and 4-tBP that were found to have a 20-fold lower rate of oxidation by
tyrosinase/O2 metabolizing system.
Tyrosinase mediated GSH depletion
assay
As shown in
Table 1, alkoxyphenols depleted GSH in the tyrosinse/O2 metabolizing
system in the following decreasing order: 4-HA >> 4-nBP, 4-iBP, and 4-nHP
>> 4-EP and 4-nPP >> 4-iPP, 2-iPP, 4-sBP and 4-tBP. 4-HA depleted
1.8 molecular equivalent of GSH per mole at 30 min incubation. 4-nBP, 4-iBP,
and 4nHP depleted 1.8 molecular equivalent of GSH per mole whereas 4-EP and
4-nPP depleted GSH on mole per mole basis after 90 min incubation. 4-iPP,
2-iPP, 4-sBP and 4-tBP depleted GSH only to an average equivalent of 0.15 molar
per mole with tyrosinase/O2 metabolizing system during the 90 min
incubation. It was found that additional substitution/s on the alpha carbon of
alkoxy group prevented the metabolism of the molecule by tyrosinase/O2.
For instance, the rate of metabolism for 4-butoxyphenol series (4-nBP, 4-iBP, 4-sBP,
and 4-tBP) by tyrosinase was diminished in the following decreasing order 4-tBP
< 4-sBP < 4-nBP and 4-iBP < 4-HA which suggests that the presence of a
non linear side chain on the alpha carbon atom of the alkoxy group may
interfere with the molecular fit into the tyrosinase enzyme active site.
Negligible GSH depletion occurred in the absence of the enzyme.
Glutathione conjugate formation by
rat hepatocyte microsomes
The amount of
GSH depleted as a result of alkoxyphenol metabolism catalyzed by rat liver
microsomes/NADPH/O2 was determined to be between 1.2 -1.5
equivalents of GSH per mole except for 4-tBP which was 0.5 equivalent per mole
of GSH after 90 min of incubation. At 100 mM concentration (Table 1), 4-tBP showed the lowest
rate of GSH depletion by microsomal P450/NADPH/O2 metabolizing system,
which corresponded to 2.3 fold less than 4-HA. This suggests that 100% of the alkoxyphenols
in the reaction mixture underwent glutathione conjugation except 4-tBP which
could be due to hindrance imposed by the presence of the bulky t-butoxy group
in its molecular structure. It was noted that the alkoxyphenols were metabolized
by CYP 2E1 induced rat liver microsomes more readily than when non-induced/standard
rat microsomal P450 system was used. Negligible GSH depletion occurred in the
absence of the enzyme.
Cell viability
Cell viability of the cultured murine B16-F0 melanoma
cell line was measured using trypan blue exclusion test (18) and was always
greater than 95% before seeding the cells into the 96 well plates for MTT
assay.
Cytotoxicity in B16-F0 melanoma cell
line
The LC50
(2 day) concentrations were determined by MTT assay (17) as a measure of melanoma
cell viability (Figure 3). The required concentrations of compounds that can cause
50% decrease in melanoma cell viability (LC50 mM) on the second day are given in Table 1. ANOVA
and regression analysis of the toxicity of each of the alkoxyphenols at various
doses showed the cytotoxicity to be dose- and time-dependent with a ranking
order of 4-nHP>>4-nPP, 4-HA, 4-sBP, 4-iBP, 4-EP>4-iPP>>4-tBP>2-iPP
except for 2-iPP.
Partition coefficient
The partition coefficients of the alkoxyphenols were estimated
using the LogP software (www.logp.com) and were in a decreasing order of
4-nHP>>4-nBP, 4-iBP, 4-s-BP, 4-tBP>>4-nPP>4-i-PP and
2-iPP>>4-EP>>4-HA, thus indicating that the lipid solubility of the
alkoxyphenol increases as the size of the aliphatic group and/or the number of
carbon atoms on the aliphatic side chain group increases (Table 1).
One parameter quantitative structure
toxicity relationship
The data in Table 1 was used to derive Eq. 1 as a
one-parameter model for quantitative structure toxicity relationship (QSTR) for
describing the toxicity of the alkoxyphenols towards B16-F0 melanoma cell line.
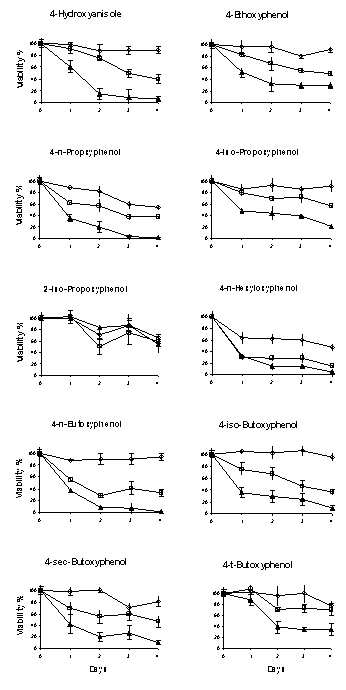
Figure
3: The cytotoxicity of alkoxyphenols towards B16-F0 mouse melanoma cell line. All assays were carried out in triplicate.
The concentrations of the reagents tested were 10 mM, 50 mM and 100
mM except for 2-iso-propoxyphenol and
4-t-butoxyphenol that were 10 mM, 100 mM and 250 mM. All the alkoxyphenols showed a dose- and
time-dependent toxicity towards B16-F0 cells except 2-iso-propoxyphenol.
4-n-Hexyloxyphenol demonstrated the greatest toxicity towards B16-F0 cells.
LogLC50 (mM)= – 0.182 (±0.153) LogP + 2.345 (±0.405) (n=10,
R2=0.150, P value for LogP term=0.268; P value for intercept
term<0.001)
Eq. 1
As
shown in Eq. 2 (Figure 4) the exclusion of outliers (4-HA, 2-iPP and 4-tBP; shown
as ● symbol) from the rest of the data points (shown as ◊ symbol)
greatly improved the QSTR equation between the alkoxyphenols LogLC50
(mM) and their LogP values. The calculated LogLC50
values from Eq. 2 were similar to the experimental data (Table 1). The outlier
4-HA (Table 1) was 1.7 fold more toxic than the toxic value calculated from Eq.
2 whereas both 2-iPP and 4-tBP were 3.6 fold less toxic. However, the toxicity
of the seven alkoxyphenols was well predicted by Eq. 2 (Table 1).
LogLC50
(mM)= – 0.265 (±0.064) LogP + 2.482 (±0.179)
(n=7, R2=0.773, P value for LogP term=0.009; P value for intercept
term<0.0001)
Eq. 2
Outliers: 4-HA, 2-iPP, and 4-tBP
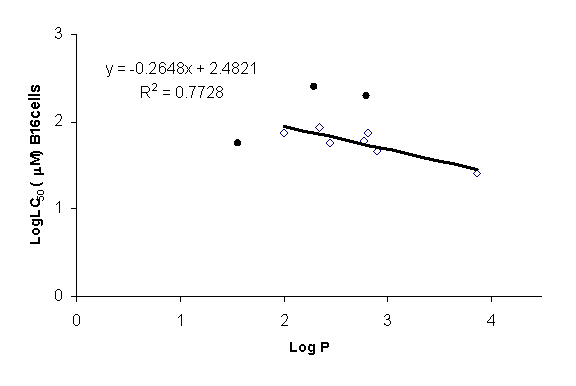
Figure 4: Graphical presentation of
quantitative structure toxicity relationship (QSTR) for alkoxyphenols in B16-F0
mouse melanoma cell line. 4-HA (4-hydorxyanisole); 4-EP
(4-ethoxyphenol); 4-nPP (4-n-propoxyphenol); 4-iPP (4-iso-propoxyphenol); 2-iPP
(2-iso-propoxyphenol); 4-nBP (4-n-butoxyphenol); 4-iBP (4-iso-butoxyphenol);
4-sBP (4-sec-butoxyphenol); 4-tBP (4-t-butoxyphenol); and 4-nHP
(4-n-hexyloxyphenol). The toxicity of alkoxyphenols increases with an increase
in its LogP value and lipid solubility. 4-n-Hexyloxyphenol demonstrated the
greatest toxicity towards B16-F0 cells. 4-HA, 2-iPP and 4-tBP are outliers as (
● ).
DISCUSSION
The aim of this investigation was to identify alkoxyphenolic
agents that were metabolized at a lower rate by rat liver microsomes/NADPH/O2
but could still be metabolized by the tyrosinase/O2 enzyme and
therefore be relatively toxic towards B16-F0 melanoma cells when compared to 4-HA.
Riley group had previously investigated the structure
activity relationship of tyrosinase dependent cytotoxicity of a series of
substituted linear alkoxyphenols (11). However, this study did not investigate
the metabolism of this group of phenols by liver P450 enzymes. Moreover, the
study did not look into the effect of substitution on the alpha and beta
carbons of the alkoxy group on the metabolism by tyrosinase/O2 and
P450/NADPH/O2 metabolizing systems. Such information is invaluable
in drug design of a safer anti-melanoma phenolic agent.
Therefore,
ten alkoxyphenol compounds (Figure 2) with various linear and branched alkoxy
side chains were selected to investigate the effect of substitution on the
alpha and beta positions of the alkoxy side chain groups on the metabolism by
tyrosinase/O2, rat liver microsomes/NADPH/O2 systems and
the toxicity towards murine melanoma B16-F0 cell line. 4-HA (4-hydorxyanisole);
4-EP (4-ethoxyphenol); 4-nPP (4-n-propoxyphenol); 2-iPP (2-iso-propoxyphenol);
4-nBP (4-n-butoxyphenol); 4-tBP (4-t-butoxyphenol); and 4-nHP
(4-n-hexyloxyphenol) were available commercially. 2-iPP was selected because it
possessed a bulky group at the ortho position to the phenolic functional group.
We hypothesized that the presence of a bulky group such as isopropoxy at ortho
position of 2-iPP would prevent the phenol group from undergoing metabolism by
tyrosinase/O2. Three additional alkoxyphenols 4-iPP
(4-iso-propoxyphenol), 4-iBP (4-iso-butoxyphenol) and 4-sBP
(4-sec-butoxyphenol) were synthesized (11). These three compounds were selected
to test if the substitution on the alpha carbon group (e.g. 4-iPP, 4-sBP and
4-tBP) could substantially reduce the metabolism by the P450 metabolizing
system without any significant change in their metabolism by the tyrosinase/O2
metabolizing system. It was hypothesized that the presence of a bulky groups
such as t-butoxy as substitutes to the methyl group of 4-HA in a phenolic compound
such as 4-tBP should lower the rate and extent of its metabolism by rat liver
P450 enzyme, largely due to restrict hindrance imposed by the size of the group
on the aromatic ring leading to a reduced rate of arene epoxide formation
(Figure 5). Such molecular optimization may also increase toxicity towards the
melanoma cell line due to the greater lipophilicity of the t-butoxy group in
comparison to the methoxy group of 4-HA.
It was noted that the replacement of each
hydrogen atom on the methoxy group (O– CH3) of 4-HA substantially
reduced the molecule’s ability to undergo metabolism by tyrosinase
monooxigenase system. For instance, 4-EP (O-CH2CH3) and
4-nPP (O-CH2CH2CH3) with only one substitution
on the alpha carbon (Figure 2) showed at least a 5-fold decrease in their
ability to undergo metabolism by tyrosinase/O2. This decrease was
compensated as the length of the aliphatic chain was increased in compounds
such as 4-nBP (O–CH2CH2CH2CH3,
4-iBP, and 4-nHP (O–CH2CH2CH2CH2CH2CH3)
which could be due to an increase in the lipophilic properties of the
corresponding molecules.

Figure
5: The proposed effect of restrict hindrance of t-butoxy on metabolism of
4-t-butoxyphenol by liver P450 enzyme and melanoma tyrosinase
The decrease in the rate of metabolism and
the extent of GSH depletion was even more significant for the alkoxyphenols having
two or three alkyl substitutes on the alpha carbon atom such as for 4-iPP
(O–CH(CH3)2), 2-iPP (O–CH(CH3)2),
4-sBP (O–CH(CH3)CH2CH3) and 4-tBP (O–C(CH3)3)
which showed a 30-fold decrease in the extent of GSH depletion by the
tyrosinase/O2 metabolizing system. For 4-tBP, this could be because it
possessed a bulkier group than other alkoxy groups. 2-iPP also possesses a
bulky group [iso-propxy (O–CH(CH3)2)] at the ortho
position to the phenolic functional group which prevents the phenol group
undergoing metabolism by tyrosinase/O2 system.
Unlike
alpha substitution, the derivatization on the beta carbon atom of aliphatic
side chain did not significantly alter the rate of the metabolism of these
compounds by tyrosinse/O2 metabolizing system (Figure 2; Table 1).
The mechanism of the 4-HA metabolism
by isolated rat hepatocytes and rat liver microsomes was previously
investigated (6). It was found that P450 plays a major role in the metabolism
of 4-HA to p-quinone and its induced cytotoxicity towards isolated rat
hepatocytes. In addition, it was shown that P450 inhibitors could significantly
abolish P450 mediated 4-HA induced cytotoxicity metabolism. Three mechanistic
pathways were proposed for 4-HA metabolism by the P450 system (6) which
included: ipso attack, O-demethylation, and arene epoxidation pathways. Because
of the absence of any electronic withdrawing group in the molecular structure
of 4-HA the ipso attack mechanism was not considered as a viable route for 4-HA
metabolism. The investigators were unable to identify formaldehyde, a metabolic
product of 4-HA metabolism if it was metabolized via the O-demethaylation
pathway. Therefore, by exclusion it was concluded that arene expoxidation was
the correct mechanistic metabolism route for p-quinone formation by the P450/NADPH/O2
metabolizing system (6). Other investigators were also suggested a similar
mechanism of metabolism for arene epoxide formation (19). These findings led us
to conclude that the introduction of a bulky group such as t-butoxy into the
chemical structure of alkoxyphenol may prevent and/or limit the metabolism of
4-t-butoxyphenol by P450 system. Besides, t-butoxy lacks a hydrogen atom on the
alpha carbon immediately next to oxygen atom (–O-C(CH3)3). This makes it unlikely for the
molecule to undergo O-dealkylation and consequently may lead to a minimal
metabolism by P450/NADPH/O2 and toxicity towards liver.
It was found that all the
alkoxyphenols tested in this work showed toxicity towards murine B16-F0
melanoma cell line. The cytotoxicity of these alkoxyphenols were shown to be
dose- and time-dependent with a ranking order of 4-nHP>>4-nPP, 4-HA,
4-sBP, 4-iBP, 4-EP>4-iPP>>4-tBP>2-iPP except for 2-iPP. Our data
also showed that 2-iPP and 4-tBP were poor substrates for tyrosinase and
thereby the least toxic substances against melanoma cells (Table 1). It was
postulated that the observed enhanced toxicity could be due to their higher
degree of lipid solubility, which provides these molecules with an ability to
cross the cell membrane more readily. Such increase in lipid solubility could
ultimately compensate for the restrict hindrance imposed by the additional
derivatization on the alpha position on alkoxy group as a preventive factor for
metabolism by tyrosinase/O2 as discussed above. Based on the data
presented in the Tables 1 and 2, it is expected that 4-nHP at LC10
and LC50 concentrations leads to a lower amount of GSH depletion by
microsomal/NADPH/O2 than 4-HA and, therefore, 4-nHP was identified
as a potential lead anti-melanoma agent in this study.
Furthermore, our data demonstrate a direct
relationship between toxicity toward B16-F0 cells and the degree of
lipophilicity via one parameter QSTR Eq. 2, which can be considered an
invaluable tool for estimating the toxicity of untested alkoxyphenols in
future. There is no clear reason for the anomalous behavior of 4-HA, 2-iPP and
4-tBP except that the alkoxy group of the latter two phenols may hinder
tyrosinase to hydroxylate the aromatic ring effectively to the corresponding
catechol analogues.
The presence of 4-HA as an outlier with
more toxicity implies that other factors or specific mechanisms other than
hydroxylation and o-quinone formation mediated by tyrosinase/O2 are
involved in the toxicity towards B16 melanoma cells. Previously Passi et al (8)
reported that melanoma toxicity might also result from inhibition of
mitochondrial electron transport. It was recently shown that polyphenols
induced hepatocyte cytotoxicity correlated with mitochondrial membrane
potential (20) and a collapse of hepatocyte mitochondrial membrane preceded the
cytotoxicity of most phenols towards rat liver hepatocytes.
Table 2: The calculated
percentage GSH depletion by microsomal metabolizing system calculated for LC10
and LC50 (mM)
of the alkoxyphenols
|
|
Lethal concentration
|
%GSH depleted by P450/NADPH/O2
metabolizing system
|
|
Alkoxyphenol
|
Calculated
LC10 (mM)a
|
Calculated
LC50 (mM)a
|
Calculated for
LC10 (mM)
concentration b
|
Calculated for
LC50 (mM)
concentration b
|
Measured at
100 (mM)
concentration
|
|
4-hydroxyanisole*
|
10**
|
58**
|
6%**
|
33%**
|
58±2**
|
|
4-n-propoxyphenol
|
1
|
58
|
<1%
|
35%
|
61±5
|
|
4-n-butoxyphenol
|
6
|
46
|
4%
|
32%
|
69±6
|
|
4-t-butoxyphenol
|
27
|
200
|
7%
|
50%
|
25±3*
|
|
4‑n‑hexyloxyphenol*
|
<1*
|
26*
|
<1%*
|
17%*
|
66±3
|
* Significantly different (t-test, P<0.05) from 4-HA
(marked as **) for selected data.
a LC10 and LC50
concentrations were calculated from equations presented in Table 1.
b GSH% depletions at LC10 and LC50
were calculated by dividing the LC concentration by 100 followed by multiplying
the product by GSH% depletion at 100 (mM).
In addition, we investigated
the QSTR for phenols (21), catechols (22) and polyphenols (23) in isolated rat
hepatocytes in which it was found that the phenols with higher lipophilicity,
bond dissociation or s+ values
or with lower pKa and redox potential were more toxic towards hepatocytes
(21-23). However, one should note that all the phenols studied in this work
differ only significantly in their lipid solubility properties but not other
physico-chemical properties. Our previous studies in hepatocyte (21-23)
indicate that one or a combination of mechanisms; i.e. mitochondrial
uncoupling, phenoxy radical, or phenol metabolism to quinone methides and
quinones, contribute to phenol cytotoxicity towards hepatocytes depending on
the phenol chemical structure. Therefore, a similar cytotoxic mechanism may
contribute to the cytotoxicity of alkoxyphenols towards melanoma B16-F0 cell
line.
In
summary, 4-nHP was identified as a potential lead anti-melanoma compound.
However, before a conclusion can be made on how effective 4-nHP might be in the
treatment of melanoma, further investigations into its mechanism of toxicity,
in vivo metabolism and pharmacokinetic profiles are required and these are
currently under investigation in our laboratory.
ACKNOWLEDGMENTS
The first author
wishes to thank School of Pharmacy,
Texas Tech University Health
Sciences Center, for an internal grant to support this research.
REFERENCES
-
Ries, L.A.G., Eisner, M.P., Kosary,
C.L., Hankey, B.F., Miller, B.A., Clegg, L., Mariotto, A., Feuer, E.J., and
Edwards, B.K. (eds).
SEER Cancer
Statistics Review, 1975-2001, National Cancer Institute.
Bethesda,
MD,
http://seer.cancer.gov/csr/1975_2001/, 2004.
-
Anderson, C.M., Buzaid, A.C., and Legha, S.S.
Systemic treatments for advanced cutaneous melanoma. Oncology, 9:1149–1158,
1995.
-
Riley, P.A. Hydroxyanisole
depigmentation: in vitro studies.
J
Pathol 1969;
97:193–206.
-
Naish, S., Cooksey, C.J., and Riley, P.A. Initial
mushroom tyrosinase-catalyzed oxidation product of 4-hydroxyanisole is
4-methoxyorthobenzoquinone.
Pigment
Cell Res,
1:379–381,
1988.
-
Naish, S., Holden, J.L., Cooksey, C.J., and Riley, P.A.
Major primary cytotoxic product of 4-hydroxyanisole oxidation by mushroom
tyrosinase is 4-methoxyorthobenzoquinone.
Pigment
Cell Res,
1:382–385,
1988.
-
Moridani,
M.Y., Cheon, S.S., Khan, S., and O'Brien, P.J. Metabolic activation of
4-hydroxyanisole by isolated rat hepatocytes. Drug Metab Dispos, 30:1063–1069,
2002.
-
Land, E.J., Cooksey, C.J., and Riley, P.A. Reaction
kinetics of 4-methoxy ortho benzoquinone in relation to its mechanism of
cytotoxicity: a pulse radiolysis study.
Biochem
Pharmacol, 39:1133–1135,
1990.
-
Passi, S., Picardo, M., and Nazzaro-Porro, M., Effect on
para-hydroxyanisole of tyrosinase and mitochondrial oxido-reductases, in Riley
PA (eds), Hydroxyanisole: Recent
Advances in Anti-Melanoma
Therapy.
IRL Press Limited,
England,
pp57–70, 1984.
-
Morgan, B.D.G., Recent results of a
clinical pilot study of intra-arterial 4-hydroxyanisole chemotherapy in
malignant melanoma, in Riley PA (eds),
Hydroxyanisole: Recent Advances in Anti-Melanoma
Therapy.
IRL Press Limited,
England,
pp
233–241,
1984.
-
Rustin, G.J.,
Stratford, M.R., Lamont, A., Bleehen, N.,
Philip, P.S., Howells, N., Watfa, R.R., and Slack, J.A., Phase 1 study of
intravenous 4-hydroxyanisole.
Eur J
Cancer,
28A:1362–1364,
1992.
-
Naish-Byfield,
S., Cooksey, C.J., Latter, A.M., Johnson, C.I., and Riley, P.A., In vitro
assessment of the structure-activity relationship of tyrosinase-dependent
cytotoxicity of a series of substituted phenols. Melanoma Res, 1:273-287, 1991.
-
Gergel,
D., and Cederbaum, A.I., Interaction of nitric oxide with 2-thio-5-nitrobenzoic
acid: implications for the determination of free sulfhydryl groups by Ellman’s
reagent.
Arch Biochem Biophys,
347:282-288,
1997.
-
Ellman, G.L., Tissue sulfhydryl
groups.
Arch Biochem Biophys,
82:70–77,
1959.
-
Dallner, G., Isolation of microsomal
subfractions by use of density gradients.
Methods Enzymol,
52:71–82,
1978.
-
Lowry,
O.H.,
Rosebrough,
N.J., Farr, A.L., and Randall, R.J., Protein
measurement with the Folin phenol reagent. J Biol Chem, 193:265-275, 1951.
-
Krikun,
G., and Cederbaum, A.I., Increased microsomal oxidation of alcohols after
pyrazole treatment and its similarities to the induction by ethanol
consumption. Biochim Biophys Acta, 801:131-137, 1984.
-
Wu,
X., Zeng, H., Zhang, X., Zhao, Y., Sha, H., Ge, X., Zhang, M., Gao, X., and
Xu, Q., Phosphatase of regenerating liver-3 promotes
motility and metastasis of mouse melanoma cells. Am J Pathol, 164:2039-2054,
2004.
-
Moridani,
M.Y., Cheon, S.S., Khan, S., and O'Brien, P.J., Metabolic activation of
3-hydroxyanisole by isolated rat hepatocytes. Chem Biol Interact, 142:317-333,
2003.
-
Guengerich,
F.P., Common and uncommon cytochrome P450 reactions related to metabolism and
chemical toxicity. Chem Res Toxicol, 14:611-650, 2001.
-
Galati, G., Teng, S.,
Moridani, M.Y., Chan, T.S., and O'Brien, P.J., Cancer chemoprevention and
apoptosis mechanisms induced by dietary polyphenolics. Drug Metabol Drug
Interact, 17:311-349, 2000.
-
Moridani,
M.Y., Siraki, A., and O'Brien, P.J., Quantitative structure toxicity
relationships for phenols in isolated rat hepatocytes. Chem Biol Interact,
145:213-223, 2003.
-
Moridani,
M.Y., Siraki, A., and Chevaldina, T., Scobie, H., and O'Brien, P.J.,
Quantitative structure toxicity relationships for catechols in isolated rat
hepatocytes. Chem Biol Interact, 147:297-307, 2004.
-
Moridani, M.Y.,
Galati,
G., and O'Brien, P.J., Comparative quantitative structure toxicity
relationships for flavonoids evaluated in isolated rat hepatocytes and HeLa
tumor cells. Chem Biol Interact, 139:251-264, 2002
JPPS
Contents
Published by the Canadian Society for Pharmaceutical Sciences.
Copyright © 1998 by the Canadian Society for Pharmaceutical
Sciences.
http://www.cspscanada.org
CSPS Home |
JPPS
Home |
Search
|
Subscribe to JPPS




