J Pharm Pharmaceut Sci (www.ualberta.ca/~csps) 6(3):321-326, 2003
Preparation and in vitro transfection efficiency of chitosan microspheres containing plasmid DNA:poly(L-lysine) complexes.
Cenk Aral1, Julide Akbuga
Marmara University, Faculty of Pharmacy, Department of Pharmaceutical Biotechnology, Haydarpasa, Istanbul, TurkeyReceived 17 June 2003, Revised 11 October 2003, Accepted 3 November 2003
PDF version
Abstract
PURPOSE: Studies on DNA complexes with cationic polymers are prompted by the search for nonviral DNA carriers for gene therapy. Among them, poly(L-Lysine) (PLL) has been extensively studied. On the other hand, these systems deliver DNA as a bolus without long-term release. The aims of this study were to encapsulate plasmid DNA:poly(L-lysine) (pDNA:PLL) complexes into chitosan microspheres as an alternative to the PLL based gene delivery and investigate its in vitro release and transfection characteristics as well as plasmid DNA integrity and stability against serum and DNase I challange. Methods. pUC18 plasmid DNA that encoded b-galactosidase was used as a model. The microspheres were prepared by complex coacervation method and the release and in vitro transfection properties were investigated. pDNA:PLL complexes were prepared at two different mass ratios. In vitro release studies were performed at 37 ± 0.5°C and drug release was monitored both spectrophotometrically and fluorometrically. Structural integrity of the pDNA:PLL complexes were determined by Southern blotting analysis. Protective effect of encapsulation of pDNA:PLL complexes against DNase I and serum treatment were also studied. In vitro transfection studies were performed by using 3T3 cell line. Results. According to our in vitro release data, the mass ratio of pDNA:PLL significantly affected the release of pDNA:PLL complexes from chitosan microspheres, and the structure of the plasmid DNA did not change during the experiments. pDNA:PLL-loaded chitosan microspheres indicated high stability against fetal bovine serum and DNase I treatment for a week. In vitro transfection data showed that pDNA:PLL-loaded chitosan microspheres could be effectively transfected 3T3 cells in vitro. Conclusion. As a conclusion, pDNA:PLL complexes could be encapsulated into chitosan microspheres with maintaining their structural and functional integrity and this system may be a good alternative for polycation based gene carriers.
Introduction
Gene therapy is a powerful treatment option for curing congenital and acquired diseases which based on the transfer of genes to the patients. Basically, there are two types of gene carriers to deliver oligonucleotides to the target cells or tissues, namely viral and non-viral vectors. Although viral vectors has capable of maintain stable and high gene expression, usefulness of these vectors is problematic due to safety concerns such as random recombination and immunogenicity (1). Besides of these major drawbacks, production of viral vectors as pharmacological products can not easily meet with the strict Good Manufacturing Practice guidelines. Thus researchers have focused on non-viral vectors. One of the non-viral approaches is the use of polycationic polymers that condensing negatively charged DNA into nanomeric particles. Among them poly(L-lysine) (PLL) has been extensively studied for gene and oligonucleotide delivery due to several advantages; i) Easily available and cost effective; ii) solubility in body fluids; iii) presence of side substituents for the ligand or drug incorporation (2-4). On the other hand, these systems deliver DNA as a bolus without long term release that results in shorter period of gene expression and repeated admistrations are necessary (5).
Another attractive way to deliver DNA and oligonucleotides to the target cells is encapsulation of them into the controlled-release systems. For this purpose, poly(lactide-co-glycolide) (PLGA) (6) and gelatin (7) microspheres have been used. Recently chitosan microspheres have been shown great promise for gene delivery (8-10). Chitosan is a polycationic, biodegradable natural polymer with low toxicity. Enhanced stability of plasmid DNA encapsulated in chitosan microspheres has also been reported (9,11). In our previous study, sustained DNA release and long term in vivo gene expression were obtained with chitosan microspheres (9,10,12).
Encapsulation of pDNA:PLL complexes into controlled release systems may be a good approach for improving the polyplex based gene delivery. Capan et al. (13,14) and Gebrekidan et al. (15) suggested the encapsulation of pDNA:PLL complexes into PLGA microspheres. However, there are some drawbacks of PLGA microspheres such as low plasmid DNA incorporation, formation of acidic products in the course of in situ polymer degradation that is degrading the plasmid DNA (6,16).
The goal of this study is to encapsulate PLL-complexed plasmid DNA into the chitosan microspheres in order to achieve long-term release as an alternative to polyplex based gene delivery. For this purpose, pDNA:PLL complexes were prepared and encapsulated into chitosan microspheres. Stability of plasmid DNA against serum and DNase I, in vitro release and cell transfection efficiency of these microspheres were studied.
Materials And METHODS
Materials
Poly(L-lysine) (PLL) (Mw: ca. 37 600 D) (Sigma, USA), chitosan (Mw: ca. 400 kD, viscosity: ca. 200 mPa in 1% acetic acid at 20°C) (Fluka, Germany), sodium sulphate (Carlo Erba, Italy), o-nitrophenil-b-D-galactopyranoside (ONPG) (ICN, USA) were used. Digoxigenin (DIG) labeling kit was obtained from Roche, Germany. All of the cell culture media and reagents were purchased from Gibco, USA. Other chemicals were of pharmaceutical or molecular grade.
Plasmid Construction
pUC18 (2.69 kbp) (Fermentas, Lithuania) that encoded b -galactosidase was used in this study. The plasmid DNA was amplified in HB101 strain of E. Coli and extracted by alkaline-lysis method according to Birnboim (17). The purity and concentration of the isolated plasmid DNA were spectrophotometrically assayed at 260 nm (Shimadzu, Japan) according to Sambrook et al (18). Purity of the isolated plasmid was also checked with agarose gel electrophoresis with ethidium bromide staining (0.7% agarose in tris-boric acid-EDTA buffer, pH 8.0).
Preparation of pDNA:PLL Complexes
Two mass ratios of pDNA:PLL complexes were prepared. 180 m g or 90 m g of PLL [solubilized in tris-EDTA (TE) buffer, pH 8.0] was added to 500 m g of plasmid DNA solution in TE buffer in order to achieve 1:0.36 and 1:0.18 mass ratios respectively. Then the mixture was rapidly vortexed and left at room temperature (25 ± 0.5°C) for 30 min. to complete complexation. The formation of the complexes was confirmed by 0.7 % agarose gel electrophoresis. Dye exclusion assay was also performed fluorometrically using Hoechst 33258 dye to further confirm the complexation. For this purpose, fluorescence emitted by plasmid DNA and Hoechst 33258 was recorded by a fluorometer (BioRad, USA) after and before PLL solution addition.
Preparation of Chitosan Microspheres Containing pDNA:PLL Complexes
Chitosan microspheres containing pDNA:PLL complexes were prepared according to earlier reports (19,10). Briefly, complexes were added to the sodium sulphate solution (20% w/v) before dropping, and this mixture was dropped into the chitosan solution (0.35 % w/v in 1% tween 80 and 2 % acetic acid) and stirred (Ika-Werk, Germany) for one hour at 500 rpm. Formed microspheres were washed with deionized water and separated by centrifugation at 11000 g (Sigma 30K, USA) then freeze-dried (Leybold-Lyovac GT2, Germany). Microspheres were stored at 4°C. Size of the microspheres was determined using an ocular micrometer on a light microscope (Olympus BH, Japan) (n=1000 particles). Encapsulation efficiency of chitosan microspheres was estimated by measuring the amount of drug in the supernatant after centrifugation (10).
In Vitro Release Studies
Release profiles of the chitosan microspheres were determined by incubating 10 mg samples of microspheres in phosphate buffered saline (PBS, pH 7.4, BP) at 37°C. Samples were removed and centrifuged for 10 min at 16000 g (Hettich, Germany). The release medium was replaced with fresh buffer after each sampling. Release studies were continued up to 40 days. All samples were measured both spectrophotometrically at 260 nm (Shimadzu, Japan) and fluorometrically by using 360/40 and 460/10 filter set (BioRad, USA) in order to determine released amount of pDNA:PLL complexes. For the fluorometric analysis, Hoechst 33258 dye that specifically binds to DNA was used. The mean of three determinations is given. The structural integrity of the plasmid DNA samples released from microspheres were also checked by Southern blotting analysis as mentioned below (n=3).
Analysis of Plasmid DNA Integrity
Southern blotting analysis was performed to check the integrity of the released plasmid DNA samples. Samples (5 ng equivalent weight of DNA in each lane) were applied to a 0.7 % agarose gel containing tris-boric acid-EDTA buffer (pH 8.3) and ethidium bromide (0.5 m g/ml) at constant voltage (80 V) (Horizontal gel apparatus system, Atto, Japan). After electrophoresis, the gel was denaturated, neutralized and equilibrated as described by Sambrook et al. (18). The DNA was transferred to a positively charged nylon membrane. Hybridization with digoxigenin labeled pUC18 probe was performed according to manufacturer' s instructions (Roche, Germany).
Stability of Plasmid DNA
The stability of plasmid DNA against DNase I and serum treatment was determined. DNase I solution (10 U) or fetal bovine serum (10 m l) was added to pDNA:PLL-loaded chitosan microspheres and unencapsulated pDNA:PLL. Uncomplexed and unencapsulated plasmid DNA was used as control also. After a week, the enzyme activity was stopped at 65 ± 0.5°C in a shaker bath. The microspheres were washed with sterile distilled water several times. Plasmid DNA was extracted and its stability confirmed by agarose gel elecrophoresis.
In Vitro Transfection Studies
In vitro transfection studies were performed as previously described (12) by using 3T3 cell line (ATTC, USA) as a model. The cells were normally grown in minimal essential medium supplemented with 10% fetal bovine serum, 100 mM L-glutamine and 100 mM antibiotic (penicillin, streptomycine, amphotericin B) at 37°C, 5% CO2 and they were trypsinized and seeded in 24-well plates (5x104 cells/well) and grown for 24 hours. pDNA:PLL containing chitosan microspheres were suspended in the 10 % serum-containing medium and added to the cells (100 m g microsphere/well). The cells were incubated for 19 hours then were washed with PBS three times. The cells were lysed by three freezing and thawing cycles and the b -galactosidase activity was determined according to our previous study (9) by using ONPG as a substrate of the enzyme. The ONPG solution was added to the cells and incubated at 37°C for 19 hours then the enzyme activity was spectrophotometrically assayed at 420 nm (n=6).
Statistical Analysis
All data were compared using the non-parametric Mann-Whitney Rank Sum Test. A probability (p) <0.05 was considered significant.
RESULTS And Discussion
By mixing the PLL and plasmid DNA solutions, an interpolyelectrolyte complex was formed as evidenced by agarose gel electrophoresis. As seen in figure 1, at 1:0.18 (w/w) pDNA:PLL ratio (lane 3), migration of plasmid DNA was slightly retarded probably due to an increase in the molecular weight or decrease in the net negative charge of the plasmid DNA as a result of PLL binding as previosly noticed by Kabanov and Kabanov (20) and Çapan et al (14).
Figure 1: Agarose gel electrophoresis of pDNA:PLL complexes. Lane 1: Hind III digested lambda DNA. Lane 2: pUC18 alone (control). Lane 3: 1:0.18 mass ratio of pDNA:PLL. Lane 4: 1:0.36 mass ratio of pDNA:PLL.
On the other hand, mobility of the plasmid DNA was completely hindered and remained in the gel slot by adding of more PLL to the plasmid DNA (eg. 1:0.36 pDNA:PLL mass ratio) (Fig 1, Lane 4). Furthermore, complex formation between plasmid DNA and PLL was also confirmed fluorometrically by dye exclusion assay using Hoechst 33258. Fluorescence emitted by plasmid DNA-dye solution was dramatically decreased by adding PLL (data not shown). These data were consistent with the previous studies concerning DNA:polycationic polymer complexes published elsewhere.
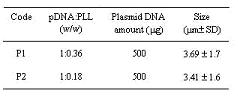
Chitosan microspheres were prepared by complex coacervation technique. pDNA:PLL complexes, prepared as 1:0.36 and 1:0.18 mass ratios, were encapsulated in to the chitosan microspheres and coded as P1 and P2 respectively (Table I).
Table 1: Codes and formulations of chitosan microspheres containing pDNA:PLL complexes.
Initial drug loading for both formulations were 0.28 % as calculated by initial plasmid DNA amount added to the formulation. Encapsulation efficiency for pDNA:PLL complexes were about 90 % (data not shown). The average sizes of chitosan microspheres were between 3.41 to 3.69 m m and independent from pDNA:PLL ratio. These findings are in accordance with our previous studies (10,12).
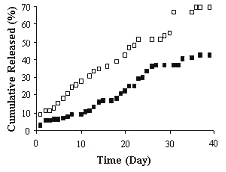
Release profiles of chitosan microspheres are given in figure 2.
Figure 2: Release profiles of chitosan microspheres containing pDNA:PLL complexes with the mass ratio of 1:0.36 (P1) (■) and 1:0.18 (P2) (❏). Mean of 3 determinations are given.
The release of pDNA:PLL complexes from the microspheres shoved no initial burst release. In the first day, 2.1 % and 4.8 % of the pDNA:PLL complexes were released from formulations P1 and P2 respectively. This was probably due to high positive charge of chitosan which could be hindered the release of surface localized plasmid DNA. Çapan et al (13), were obtained high burst release from PLGA microspheres and they concluded that was due to fast release of surface localized plasmid DNA. In vitro release studies showed that the ratio of pDNA:PLL was significantly affected the drug release from the chitosan microspheres (p<0.05). Microspheres containing pDNA:PLL complexes at the ratio of 1:0.18 (w/w) (P2) was indicated faster release than the other formulation (P1; 1:0.36 pDNA:PLL mass ratio) (Fig 2). At day 40th, 42.35 % of the initial drug was released from chitosan microspheres for formulation P1 while 69.65 % for formulation P2 (Fig 2). This may be due to lower solubility of highly complexed plasmid DNA. Gebredikan et al (15), indicated faster release of free plasmid DNA than the complexed one from PLGA microspheres and they concluded that different release patterns of free and complexed DNA was due to porous structure of the PLGA microspheres. On the other hand, it is known that cationic polymer binding to the negatively charged plasmid DNA results in increase of the molecular weight of plasmid DNA as pointed above. Luo at al (5) were showed that higher molecular weight DNA have lower diffusion constant which results in slower release rate.
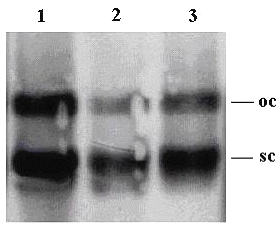
In order to determine the structural integrity of the released plasmid DNA from chitosan microspheres, southern blotting analysis was performed. As seen in figure 3, two topological forms of released plasmid DNA [namely super coiled (sc) and open circular (oc)] is appeared in the gel electrophoresis (lanes 2 and 3).
Figure 3: Southern blotting analysis of released plasmid DNA from chitosan microspheres. Lane 1: pUC18 alone (control). Lane 2 and 3: Release samples of formulations P1 and P2 respectively. (oc: open circular, sc: supercoiled forms of plasmid DNA).
Similar topological forms were also present in initial plasmid DNA (Fig 3 lane 1). It is seen that there is no additional band or an extensive smear, so the integrity of the plasmid DNA was not changed during the experiments. Although, there was no DNA migration at initial experiments of 1:0.36 pDNA:PLL complexes (Fig 1, Lane 4) two plasmid DNA bands were also appeared at the release samples of formulation P1 (Fig 3, Lane 2). This is suggested that some degree of decomplexation occurred however mechanism lying under the decomplexation was not clear.
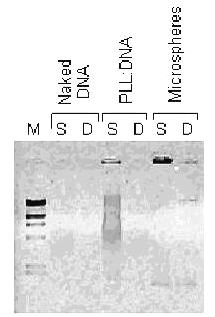
Nuclease sensitivity of naked DNA is one of the main causes of low transfection efficiency. Condensing DNA with cationic polymers improves the resistance of plasmid DNA against enzymatic digestion by altering the accessibility of the DNA to the enzymes (1). Thus enhancing the stability against enzymatic degradation has become important for successful gene therapy. According to our in vitro studies, PLL complexed-plasmid DNA could be protected for a week by encapsulation into the chitosan microspheres (Fig 4).
Figure 4: Stability against serum and DNase I treatment. S and D are indicated serum and DNase I treatment respectively. pDNA:PLL mass ratio is 0.18 and microsphere samples are used from formulation P2. M is Hind III digested lambda DNA as mentioned in fig 1.
An enhanced stability with the encapsulation of pDNA:PLL complexes into PLGA microspheres were previously reported (15). Also in our previous studies, protective effect of chitosan microspheres were noted (9,10).
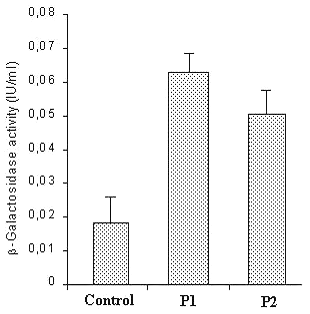
In vitro transfection studies were also peformed with pDNA:PLL-loaded chitosan microspheres. The microspheres were added to 3T3 cells and β -galactosidase activity was measured. The results are given in figure 5.
Figure 5: In vitro transfection efficiency of pDNA:PLL-loaded chitosan microspheres. Mean of 6 determinations with SD are given.
Untreated cells were taken as control group. Formulations P1 and P2 were showed a significant b-galactosidase activity (p<0.0001) (Fig 5). On the other hand pDNA:PLL ratio significantly affected the in vitro protein production and the ratio of 1:0.36 (w/w) showed higher b-galactosidase activity (p=0.0081). These data were showed that the functional integrity of the plasmid DNA was also maintained in chitosan microspheres.
In conclusion, pDNA:PLL complexes were successfully encapsulated within chitosan microspheres and achieved long-term drug release and relatively high expression in 3T3 cells. On the other hand encapsulation of pDNA:PLL complex using chitosan microspheres provides a stable delivery system against serum and DNase I challange for plasmid DNA also.
References
Smedt, S.C.D., Demeester, J. and Hennink, W.E., Cationic polymer based gene delivery systems. Pharm. Res., 17:113-126, 2000.
Wu, G.Y. and Wu, C.H., Receptor-mediated gene delivery and expression in vivo. J. Biol. Chem., 263:14621-14624, 1988.
Wagner, E., Zenke, M., Cotton, M., Beug, H. and Birnstiel, M.L., Transferrin-polycation conjugates as carriers for DNA uptake into cells. Proc. Natl. Acad. Sci. USA, 87:3410-3414, 1990.
Ferruti, P., Knobloch, S., Ranucci, E., Duncan, R. and Gianasi, E., A novel modification of poly(L-lysine) leading to a soluble cationic polymer with reduced toxicity and with potential as a transfection agent. Macromol. Chem. Phys.,199:2565-2575, 1998.
Luo, D., Mumford, K.W., Belcheva, N. and Saltzman, W.M., Controlled DNA delivery systems. Pharm. Res.,16:1300-1308, 1999.
Wang, D., Robinson, D.R., Kwon, G.S. and Samuel, J., Encapsulation of plasmid DNA in biodegradable poly(D,L-lactid-co-glycolic acid) microspheres as a novel approach for immunogene delivery. J. Cont. Rel., 57:9-18, 1999.
Truong-Le, V.L., August, J.T. and Leong, K.W., Controlled gene delivery by DNA-gelatin nanospheres. Hum. Gene Ther.,9:1709-1717, 1998.
Roy, K., Mao, H.Q. and Leong, K.W., DNA-Chitosan nanospheres: Transfection efficiency and cellular uptake. Proc. Int. Sym. Control. Rel. Bioact. Mater.,24:673-674, 1997.
Aral, C., Ozbas-Turan, S., Kabasakal, L., Keyer-Uysal, M. and Akbuga, J., Studies of plasmid DNA-loaded chitosan microspheres: I. Plasmid size, chitosan concentration and plasmid addition techniques. STP Pharm. Sci.,10:83-88, 2000.
Özbas-Turan, S., Aral, C., Kabasakal, L., Keyer-Uysal, M. and Akbuga, J., Co-encapsulation of two plasmids in chitosan microspheres as a non-viral gene delivery vehicle. J. Pharm. Pharmaceut. Sci., 6:27-32, 2003.
Mao, H.Q., Roy, K., Troung-Le, V.L., Janes, K.A., Lin, K.Y., Wang, Y., August, J.T. and Leong, K.W., Chitosan-DNA nanoparticles as gene carriers: synthesis characterization and transfection efficiency. J. Cont. Rel., 70:399-421, 2001.
Akbuga, J., Aral, C., Ozbas-Turan, S., Kabasakal, L. and Keyer-Uysal, M., Transfection efficiency of chitosan microspheres: effect of DNA topology. STP Pharm. Sci., 13:99-103, 2003.
Capan, Y., Woo, B.H., Gebrekidan, S., Ahmed, S. and DeLuca, P.P., Influence of formulation parameters on the characteristics of poly(D,L-lactide-co-glycolide) microspheres containing poly(L-lysine) complexed plasmid DNA. J. Cont. Rel., 60:279-286, 1999.
Capan, Y., Woo, B.H., Gebrekidan, S., Ahmed, S. and DeLuca, P.P., Preparation and characterization of poly(D,L-lactide-co-glycolide) microspheres for controlled release of poly(L-lysine) complexed plasmid DNA. Pharm. Res., 16:509-513, 1999.
Gebrekidan, S., Woo, B.H. and DeLuca, P.P., Formulation and in vitro transfection efficiency of poly(D,L-lactide-co-glycolide) microspheres containing plasmid DNA for gene delivery. AAPS Pharm. Sci. Tech., 1: 2000.
Walter, E., Moelling, K., Pavloviç, J., and Merkle, H.P., Microencapsulation of DNA using poly/D,L-lactic-co-glycolide) stability issues and release characteristics. J. Cont. Rel., 61: 361-374, 1999.
Birnboim, H.C., A rapid extraction method for the isolation of plasmid DNA. Meth. Enzymol., 100:243-255, 1983.
Sambrook, J., Fritsch, E.F., Maniatis, T., Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, New York, 1989.
Berthold, A., Cremer, K. and Kreuter, J., Preparation and characterization of chitosan microspheres as drug carriers for prednisolone sodium phosphate as model for anti inflammatory drugs. J. Cont. Rel., 39:17-25, 1996.
Kabanov, A.V. and Kabanov, V.A., DNA complexes with polycations for the delivery of genetic material into cells.
Corresponding Author: Cenk Aral, Marmara University, Faculty of Pharmacy, Department of Pharmaceutical Biotechnology, Tibbiye C., No.49, 81010 Haydarpasa, Istanbul, Turkey, c_aral@hotmail.com
Published by the Canadian Society for Pharmaceutical Sciences.
Copyright © 1998 by the Canadian Society for Pharmaceutical Sciences.
http://www.ualberta.ca/~csps