J Pharm Pharmaceut Sci (www.ualberta.ca/~csps) 5(3):279-284, 2002
Analysis of peptide and lipopeptide content in liposomes.
M. E. Christine Lutsiak, Glen S. Kwon, and John Samuel1
Faculty of Pharmacy and Pharmaceutical Sciences, University of Alberta, Edmonton, Alberta, Canada; School of Pharmacy, University of Wisconsin, Madison, Wisconsin, USA
Received 6 August 2002, Revised 8 November, Accepted 9 November 2002
PDF version
Abstract
PURPOSE: To evaluate several methods for extraction of peptides from liposomal formulations as a first step in their quantification, and to determine the encapsulation efficiency for a panel of 8 peptides. METHODS: Eight peptides were chosen due to their importance in the field of vaccine development. Three different extraction media were examined: 25% ethanol, 98% ethanol, and 100% methanol. After extraction from liposomes, peptide content was measured using reverse phase HPLC. RESULTS: The effectiveness of the extraction media for peptide recovery varied considerably for the different peptides studied. In general, more hydrophilic peptides were recovered to a greater extent using 25% ethanol while more hydrophobic peptides were more thoroughly recovered using 98% ethanol. Encapsulation efficiencies (EE) ranged from 1% to 99% for the different peptides. No strong correlation was found between the average hydrophobicity values for the peptides and their EE. CONCLUSIONS: The most effective solvent for the extraction of a peptide from liposomes depends on the physicochemical properties of the peptide. Although the peptide sequence characteristics may provide guidance on the choice of the extraction media, only peptide recovery experiments will be able identify the optimal medium for extraction..
Introduction
Encapsulation of peptide antigens in liposomes protects them from degradation and enhances their immunogenicity. Prior to use, vaccine formulations of peptides must be fully characterized with respect to the peptide content and physicochemical properties of the formulations. The dose of the peptide antigen can significantly influence the type and magnitude of the immune response, which may have detrimental or beneficial effects in several disease states (1, 2). Therefore, the development of methods that permit rapid and accurate determination of peptide content in particulate formulations is important. Such methods are also essential for scale up, quality control, and determination of batch-to-batch consistency of the formulations.
Quantification of an encapsulated drug may be done either without disruption of the particles or after its extraction from the formulation. Direct estimation of an encapsulated compound in a formulation may be performed based on its fluorescence properties (3, 4) or radioactivity (5-8). However, if fluorescence or radioactivity is introduced into the compound by significant chemical modification, the encapsulation efficiency of the modified compound may vary significantly from that of the native molecule of interest. In addition, the labeling process needs to be optimized for each molecule under investigation, which can be time consuming and expensive. A more effective and widely applicable method would be the quantification of the compound after its extraction from the formulation matrix. Encapsulated drugs can be extracted from liposomes by various extraction media including Triton-X (3, 9-12), chloroform (8), methanol (13), sodium chlorate (14), octyl-b-glucoside (14), sodium dodecyl sulphate (15), or acidified isopropanol (16). Once the encapsulated compound is separated from the lipids, it can be quantified in several ways, including reverse phase (RP) HPLC (9, 16).
RP-HPLC is the standard method for quantification of peptides and is favoured for the determination of peptide encapsulation efficiency in vaccine formulations for several reasons. First, RP-HPLC allows the determination of peptide content without interference by lipids or polymer. In addition, the UV absorption spectra of the encapsulated peptide and unencapsulated peptide can be compared to ensure that the peptide has not been altered by encapsulation or extraction. Finally, RP-HPLC is an easy and replicable method and is suitable for gauging relatively small amounts of peptide.
Although the extraction of the encapsulated drugs from liposomes is a widely used step in their quantification, it has not been adequately explored in the context of therapeutic peptides. Similarly, the use of RP-HPLC to determine peptide quantity is a standard procedure that has yet to be carefully examined in the framework of liposomal and polymeric formulations. The purpose of this series of investigations was to determine how best to extract a variety of peptides and lipopeptides from vaccine formulations in order to examine encapsulation efficiency using an RP-HPLC.
MATERIALS AND METHODS
Materials
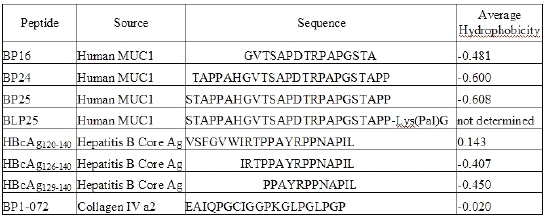
A panel of peptides of varying hydrophobicity/hydrophilicity characteristics, prepared by solid-phase synthesis, from MUC1 mucin, hepatitis B virus core antigen (HBcAg), and collagen IV were used. Amino acid sequences of the peptides are reported in Table 1.
Table 1: Peptide Sources and Amino Acid Sequences
Average hydrophobicity (AH) is the sum of all hydrophobicity values (17) for the given sequence divided by the sequence length (Table 1). The higher the AH value, the more hydrophobic the molecule. AH values were calculated using PepTools Version 2.0 (BioTools Inc., Edmonton, AB, Canada). Peptides from the hepatitis B core antigen were synthesized by Dr. David Wishart's laboratory, Faculty of Pharmacy and Pharmaceutical Sciences, University of Alberta (Edmonton, AB, Canada). Biomira Inc. (Edmonton, AB, Canada) supplied all other peptides. The lipids used were dipalmitoyl phosphatidylcholine (DPPC), dimyristoyl phosphatidylglycerol (DMPG) (Genzyme Pharmaceuticals, Cambridge, MA, USA), and cholesterol (CHOL) (Sigma Chemical Co., St. Louis, MO, USA).
Preparation of Liposomes
Liposomes were prepared using a modified freeze-thaw method (18). Briefly, lipids were used at a molar ratio of 3 DPPC: 1 CHOL: 0.25 DMPG. Lipids in solution were coated onto round-bottom flasks using a rotary evaporator (Buchi RE111 Rotavapor, Buchi Laboratiums-Technik, Switzerland). The volumes used were 1.95 ml of DPPC (16 mg/ml) in CHCl3 , 1.8 ml of CHOL (3.08 mg/ml) in CHCl3, and 2.2 ml of DMPG (1.1 mg/ml) in a mixture of methanol and CHCl3 (1:3). The flasks were incubated at 43°C in a vacuum oven (250 mmHg) overnight to remove the residual solvent. For empty liposomes, the lipids were rehydrated with 2 ml of PBS. For liposomes containing peptide, lipids were rehydrated with 2 ml of PBS containing 300 ug of the appropriate peptide. Flasks were incubated at 53°C in a water bath and then vortexed until lipids were removed from the sides of the flasks and the solution appeared homogenous. Five cycles of the following were performed: freeze flasks in dry ice/acetone, thaw for 40 minutes at room temperature, incubate at 41°C in a water bath for 5 min, vortex for 30 s. Liposomes were collected via ultracentrifugation (Model LB-55, Beckman Instruments Inc., Mississauga, ON, Canada), at 50,000 x g for 20 minutes and washed twice with PBS. Supernatants (S1 and S2) were collected after each centrifugation for analysis of peptide content. The liposome pellet was resuspended in a replacement volume of PBS.
Table 2: Elution Conditions
RP-HPLC Analysis
Samples for the analysis of peptide recovery from liposomes and encapsulation efficiency of liposomes were manually injected into a Waters 625LC HPLC (Waters, Mississauga, ON, Canada). The RP-HPLC was equipped with a C18 reverse phase column (8 x 100 mm) and an UV detector (Waters 486) set at 210 nm. The mobile phases employed were A [10% acetonitrile (Fisher Scientific, Edmonton, AB, Canada) and 0.1% trifluoroacetic acid (Sigma, St. Louis, MO, USA) in water] and B (70% acetonitrile and 0.085% trifluoroacetic acid in water). Elution conditions for each peptide are outlined in Table 2. The peptide concentration in all samples was determined by comparing the UV absorbance of the peptide with a standard curve (R2 >0.998) generated under the same conditions.
Peptide Recovery
All experiments were set up in triplicate in microcentrifuge tubes. To each tube was added 150 μl of empty liposomes. Liposomes were spun down at 10,300 x g for 12 minutes and all supernatant was removed. To the pellets, 40 μl of the appropriate peptide was added in 20 μl of PBS. Tubes were vortexed to thoroughly mix the peptides with liposomes. To each tube was added 200 μl of 100% methanol, 200 ml of 98% ethanol, or 200 μl 25% ethanol (ethanol diluted in water). The tubes were vortexed until the lipid pellets broke up. Samples were then incubated at 51°C in a water bath for 30-60 minutes. After removal from the water bath, tubes were centrifuged at 10,300 x g for 12-15 minutes. The supernatant was analyzed for peptide content by RP-HPLC; two injections of 50 μl each were made to the RP-HPLC. For each peptide/extraction media combination there were six injections, 2 each from the 3 tubes. The amount of peptide in each injection was calculated using a standard curve. Standard curves for all peptides used in this set of investigations had R2 values of >0.998. The 6 quantities were averaged and compared to the amount of peptide added to the liposomes.
%REC = PREC/PADD X 100 (1)
where %REC is the percent recovery, PREC is the peptide recovered after mixing with liposomes, and PADD is the amount of peptide added to empty liposomes.
Encapsulation Efficiency
All experiments were set up in triplicate in microcentrifuge tubes. Samples of liposome suspensions (150 ml) were added to microcentrifuge tubes, centrifuged at 10,300 x g for 12 minutes, and 100 ml of supernatant (S3) was removed from each tube. To the liposome pellets was added 150 ml of the extraction media that proved to give the best recovery for the peptide in the peptide recovery experiments. The tubes were vortexed to break up the liposome pellet and incubated at 53°C in a water bath for 30 minutes. Tubes were centrifuged at 10,300 x g for 12-15 minutes. Two injections of 50 ml aliquots of supernatant (L for liposomes) were injected into the RP-HPLC to determine the amount of peptide encapsulated in liposomes. Fifty microliters each of a 1:3 aqueous dilution of S1 (the first supernatant removed during the preparation of the liposomes) and undiluted S2 (the second supernatant removed during the preparation of the liposomes) and S3 was injected into the RP-HPLC. The quantity of peptide in the injected volumes was calculated by comparison with a standard curve (R2 >0.998) for the peptide. The quantity of the peptide in supernatants S1, S2, and S3 were calculated taking into account the dilution factor and the total volume of each supernatant. The total amount of peptide encapsulated in liposomes (Ltotal ) was calculated based on the value of peptide in L, the total volume of liposomal batch, and the % recovery efficiency of the extraction procedure. The EE and the total amount of peptide accounted for in the analysis were calculated using equations 2 and 3.
EE = LTOTAL/PTOTAL X 100 (2)
where EE is the encapsulation efficiency of the peptide in liposomes, LTOTAL is the total amount of peptide in liposomes (L + S3), and PTOTAL is the total amount of the peptide used for formulation.
%ACC = (S1 + S2 + LTOTAL)/ PTOTAL X 100 (3)
where %ACC is the percentage of peptide accounted for, S1 is the amount of peptide in the first wash, S2 is the amount of peptide in the second wash, LTOTAL is the total amount of peptide in the liposomes, and PTOTAL is the total amount of peptide used for formulation.
Results
Peptide Recovery
Preliminary experiments confirmed the solubility of the peptides used in the candidate solvents. In order to select an appropriate extraction medium, the efficiency of the extraction media for peptide recovery from liposomal formulations was determined. For this purpose, peptide-free liposomal suspensions were spiked with a known amount of peptide and the peptide was extracted using different solvents and quantified by RP-HPLC. Comparison of the peptide recoveries allowed the evaluation of the three extraction media and the selection of the optimal solvent for determination of the encapsulation efficiency of each peptide. It also supplied a recovery value, which was used as a correction factor in calculating the encapsulation efficiency.
For each solvent, there was a wide range in the recovery values seen with the different peptides (Table 3).
Table 3: Peptide Recovery from Liposomes (mean+/- SD)
For 25% ethanol, the values ranged from 10% for the lipopeptide, BLP25, to 113 % for BP1-072, a collagen IV peptide. For 98% ethanol, values were seen from 20% for BP16 to 88% for the shortest HBcAg peptide. Recovery values for extractions using 100% methanol ranged from 45% for BP16 to 87% for BP1- 072. For four peptides, BP16, BP24, BP1- 072, and BP25, recovery was clearly greatest with 25% ethanol followed by recovery with 100% methanol and then 98% ethanol.
BP25 and BLP25 have the same amino acid sequence, differing only in the addition of a lipid moiety. Not surprisingly, this lipid moiety caused a difference in the effectiveness of recovery by different solvents. The sole lipopeptide in the experiment, BLP25, was most effectively recovered using 98% ethanol, although 100% methanol was a close second. BLP25 was extremely poorly recovered using 25% ethanol.
For the longest hepatitis B core peptide, HBcAg120-140 , recovery was greatest with 98% ethanol, and there was little difference between 100% methanol and 25% ethanol. There were only small differences seen between the three solvents in the recovery of HBcAg126-140 , but 98% ethanol appears to be the best solvent. HBcAg129-140 showed a different pattern than the other two peptides from the core antigen. The best solvent for recovery of the shortest HBcAg peptide was 25% ethanol, followed by 98% ethanol, with 100% methanol being the least useful solvent.
Encapsulation Efficiency
Each peptide was encapsulated into liposomes using the freeze thaw method, commonly used for encapsulation of hydrophilic molecules. For each peptide, the extraction medium that gave the highest recovery in the recovery experiment was used to extract the peptide from the liposomes for the encapsulation experiment.
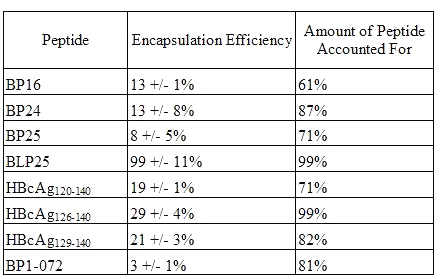
Encapsulation efficiencies of the different peptides (Table 4) ranged considerably.
Table 4: Encapsulation Efficiency of Peptide in Liposomes (mean +/- SD)
BP1-072, a collagen peptide, gave the lowest encapsulation efficiency at 3% while virtually all (99%) of the lipopeptide, BLP25, was encapsulated inside the liposomes. The peptide containing an amino acid sequence identical to that of BLP25 but lacking the lipid moiety, BP25, had an EE of only 8%. The addition of the lipid moiety to the molecule greatly enhanced the level of encapsulation of the peptide.
The total amount of peptide accounted for was considerably less than 100% in most cases. Whether this was due to peptide loss during the preparation of the liposomes or due to inefficiencies in the extraction of the peptide from the liposomes remains to be seen.
Discussion
As the number of peptide-based vaccines under investigation escalates, the development of methods to characterize peptide formulations becomes crucial. An important part of such characterization is the quantification of the encapsulated peptide antigen. Among the various methods used for determination of the EE of a peptide in a particulate delivery system, the extraction of the peptide from the formulation using a solvent, followed by the analysis of peoptide quantity by RP-HPLC, is the most advantageous for several reasons. First, it is replicable and easy to use. Secondly, it does not require chemical modification of the peptide. Finally, it can be used to analyze relatively small amounts of peptide.
Prior to using the RP-HPLC to determine the amount of peptide encapsulated inside liposomes, the peptide must be extracted from the liposomes using a solvent. The ideal solvent will be relatively inexpensive, readily available, non-toxic, and compatible with the RP-HPLC and its solutions. Many different solvents have been utilized for this purpose, but to date there has been no systematic study of which solvents are most effective for the extraction of peptides from liposomes prior to analysis of peptide content using the RP-HPLC.
In order to examine the ability of several solvents to recover peptides from liposomes, empty liposomes were spiked with a known amount of peptide and three different extraction procedures were followed to determine the recovery of eight different peptides. The results demonstrated that the amount of peptide recovered depended on the combination of extraction medium and peptide. In particular, the optimal extraction medium depended on the chemical nature of the peptide being extracted. For the more hydrophilic peptides, HBcAg129,140 , BP16, BP24, and BP25, 25% ethanol proved to be the most effective extraction medium. For the more hydrophobic peptides, BLP25 and HBcAg120-140 , 98% ethanol was the most valuable solvent.
This investigation demonstrates that the best solvent for extraction of peptide from liposomes varies, depending on the nature of the peptide being examined. It may be possible to predict the optimal solvent by comparing the amino acid sequences of an untested peptide to tested peptides or by using software capable of predicting the average hydrophobicity of a peptide, however, this can only serve as a general guideline for the selection of the extraction media. The true suitability of the solvent should be verified experimentally by determining peptide recovery from the formulation. Furthermore, in addition to the chemical nature of the peptide, the nature of the liposomal lipids may also significantly influence the optimal choice of the extraction medium. This requires further investigation.
In selecting the most effective extraction medium for a particular peptide, both the actual recovery value and the reproducibility of the recovery value must be considered. For certain recovery medium/peptide combinations, the percent coefficient of variation (% CV), an important measure of reliability, was overly high, indicating that the method was not dependable. For each peptide, an extraction medium was selected that recovered a maximal amount of peptide with a minimal amount of variation.
Once the most effective extraction medium for each peptide from liposomes was identified, the EE was ascertained for each of the eight peptides in liposomes. The results established that there was a wide range of EE for the peptides we examined. It is known that the nature of the molecule affects the incorporation of the molecule into liposomes (19) and the results from these investigations supported this. The lipopeptide gave the highest EE; its lipid moiety bestowed an amphipathic nature to the molecule that allowed for an easier encapsulation. For some of the peptides we examined, such as BP1-072, which was encapsulated at about 2% efficiency, chemical modification or encapsulation by alternate methods may be necessary to increase the EE without decreasing the antigenicity of the molecule. Interestingly, there was no correlation found between the average hydrophobicity values and the amount of peptide encapsulated inside the liposomes. It is likely that both the amino acid sequence and the distribution of hydrophilic and hydrophobic residues in the molecule play more significant roles in determining the EE of the peptides than the average hydrophobicity alone.
The peptides used in this study were selected due to their significance in the development of therapeutic vaccines. MUC1 mucin is a very important cancer antigen expressed by most carcinomas including breast, ovarian, prostate, lung, head and neck cancer and is a major target for therapeutic vaccine design (20, 21). A liposomal formulation of lipopeptide BLP25 has progressed through Phase IIb clinical trials in over 100 patients with non-small cell lung carcinoma and was shown to be safe (unpublished results). Hepatitis B virus (HBV) core antigen and its peptide epitopes are candidates for therapeutic vaccines for HBV chronic infection and hepatocellular carcinoma (22, 23). The collagen IV peptide used in this study is used as a model peptide in the development of vaccine formulations (24, 25). As more potential antigens are identified in cancer and viral disease, more peptide epitopes will be incorporated into vaccine formulations and these formulations will need to be thoroughly characterized using methods like the one described in this paper.
Acknowledgments
This work was supported by research grants from Canadian Institute of Health Research (CIHR mop 42407 and Natural Sciences and Engineering Council of Canada (NSERC stpgp 234866). C.L. was supported by an Alberta Heritage Foundation for Medical Research Studentship.
References
Bretscher, P. A., Wei, G., Menon, J. N. and Bielefedt-Ohmann, H. Establishment of stable, cell-mediated immunity the makes "susceptible" mice resistant to Leishmania major. Science, 257: 538-42, 1992.
Parish, C. R. The relationship between humoral and cell mediated immunity. Transplant. Rev., 13: 35-66, 1972.
Schroit, A. J. and Fidler, I. J. Effects of liposome structure and lipid composition on the activation of the tumouricidal properties of macrophages by liposomes containing muramyl dipeptide. Cancer Res., 42: 161-7, 1982.
Hernandez, J., Estelrich, J. and Pouplana, R. Determination of the encapsulation efficiency in liposomes obtained by the 'extruder method'. J. Microencap., 4: 315-20, 1987.
Yachi, K., Harashima, H., Kikuchi, H., Sudo, R., Yamauchi, H., Ebihara, K., Matsuo, H., Funato, K. and Kiwada, H. Biopharmaceutical evaluation of the liposomes prepared by rehydration of freeze-dried empty liposomes (FDELs) with an aqueous solution of a drug. Biopharm. & Drug Disp., 17: 589-605, 1996.
Pick, U. Liposomes with a large trapping capacity prepared by freezing and thawing of sonicated phospholipid mixtures. Arch. Biochem. Biophys., 212: 186-94, 1981.
Arien, A. and Dupuy, B. Encapsulation of calcitonin in liposomes depends on the vesicle preparation method. J. Microencap., 14: 753-60, 1997.
Alino, S. F., Garcia-Sanz, M., Irruarrizaga, A., Alfaro, J. and Hernandez, J. High encapsulation efficiencies in sized liposomes produced by extrusion of dehydration-rehydration vesicles. J. Microencap., 7: 497-503, 1990.
Turanek, J., Zaluska, D. and Neca, J. Linkup of a fast protein liquid chromatography system with a stirred thermostated cell for sterile preparation of liposomes by the proliposome-liposome method: application to encapsulation of antibiotics, synthetic peptide immunomodulators, and a photosensitizer. Anal. Biochem., 249: 131-9, 1997.
Ohsawa, T., Miura, H. and Harada, K. A novel method for preparing liposome with a high capacity to encapsulate proteinous drugs: freeze-drying method. Chem. Pharm. Bull., 32: 2442-5, 1984.
Ohsawa, T., Miura, H. and Harada, K. Evaluation of a new liposome preparation technique, the freeze-thawing method, using L-asparaginase as a model drug. Chem. Pharm. Bull., 33: 2916-23, 1985.
Ohsawa, T., Miura, H. and Harada, K. Improvement of encapsulation efficiency of water-soluble drugs in liposomes formed by the freeze-thawing method. Chem. Pharm. Bull., 33: 3945-52, 1985.
Shibata, A., Nakajima, K., Ueno, S. and Yamashita, T. Preparation and characterization of liposomes incorporating hydrophobic poly(amino acid)s with different secondary structure. Chem. Pharm. Bull., 42: 1151-3, 1994.
Liu, L. and Yonetani, T. Preparation and characterization of liposome-encapsulated haemoglobin by a freeze-thaw method. J. Microencap., 11: 409-21, 1994.
Panico, A., Pignatello, R., Cardile, V. and Puglisi, G. Preparation of liposome formulations containing immunomodulatory peptides. Pharm. Acta Hel., 72: 1-10, 1997.
Ye, Q., Asherman, J., Stevenson, M., Brownson, E. and Katre, N. V. DepoFoam technology: a vehicle for controlled delivery of protein and peptide drugs. J. Control. Release, 64: 155-66, 2000.
Kyte, J. and Doolittle, R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol., 157: 105-32, 1682.
Guan, H. G., Budzynski, W., Koganty, R. R., Krantz, M. J., Reddish, M. A., Rogers, J. A., Longenecker, B. M. and Samuel, J. Liposomal formulations of synthetic MUC1 peptides: Effects of encapsulation versus surface display of peptides on immune responses. Bioconjug. Chem., 9: 451-8, 1698.
Kulkarni, S. B., Betageri, G. V. and Singh, M. Factors affecting microencapsulation of drugs in liposomes. J. Microencap., 12: 229-46, 1695.
Samuel, J. and Longenecker, B. M. Development of active specific immunotherapeutic agents based on cancer-associated mucins. Pharm. Biotechnol., 6: 875-90, 1695.
Taylor-Papadimitriou, J., Burchell, J., Miles, D. W. and Dalziel, M. MUC1 and cancer. Biochim. Biophys. Acta, 1455: 301-10, 1699.
Schodel, F. Hepatitis B virus core and e antigen: immune recognition and use as a vaccine carrier moiety. Intervirology, 39: 104-10, 1696.
Michel, M. L., Pol, S., Brechot, C. and Tiollais, P. Immunotherapy of chronic hepatitis B by anti HBV vaccine: from present to future. Vaccine, 16: 2395-9, 2001.
Constant, S. L. and Bottomly, K. Induction of Th1 and Th2 CD4+ T cell responses: the alternative approaches. Annu. Rev. Immunol., 15: 297-322, 1697.
Murray, J. S., Madri, J., Pasqualini, T. and Bottomly, K. Functional CD4 T cell subset interplay in an intact immune system. J. Immunol., 150: 4270-6, 1693.
Corresponding Author: John Samuel, 3118 Dentistry/Pharmacy Centre, University of Alberta, Edmonton, Alberta, Canada, T6G 2N9. jsamuel@pharmacy.ualberta.ca
Published by the Canadian Society for Pharmaceutical Sciences.
Copyright © 1998 by the Canadian Society for Pharmaceutical Sciences.
http://www.ualberta.ca/~csps